Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocrosslinking
- PMID: 21486563
- PMCID: PMC3109430
- DOI: 10.1016/j.febslet.2011.04.009
Recognition of an ERAD-L substrate analyzed by site-specific in vivo photocrosslinking
Abstract
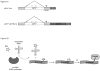
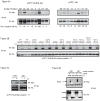
Misfolded, luminal endoplasmic reticulum (ER) proteins must be recognized before being degraded by a process called ERAD-L. Using site-specific photocrosslinking in Saccharomyces cerevisiae, we tested luminal interactions of a glycosylated ERAD-L substrate with potential recognition components. Major interactions were observed with Hrd3p. These are independent of the glycan and of other ERAD components, and can occur throughout the length of the unfolded substrate. The lectin Yos9p only interacts with a polypeptide segment distant from the degradation signal. Hrd3p may thus be the first substrate-recognizing component. Der1p appears to have a role in a pathway that is parallel to that involving Hrd3p.
Copyright © 2011 Federation of European Biochemical Societies. All rights reserved.
Figures


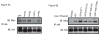
References
-
- Bagola K, Mehnert M, Jarosch E, Sommer T. Protein dislocation from the ER. Biochim Biophys Acta. 2011:925–936. - PubMed
-
- Xie W, Ng DTW. ERAD substrate recognition in budding yeast. Semin Cell Dev Biol. 2010:533–539. - PubMed
-
- Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006:361–373. - PubMed
-
- Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004:38369–38378. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

