cn.FARMS: a latent variable model to detect copy number variations in microarray data with a low false discovery rate
- PMID: 21486749
- PMCID: PMC3130288
- DOI: 10.1093/nar/gkr197
cn.FARMS: a latent variable model to detect copy number variations in microarray data with a low false discovery rate
Abstract
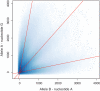
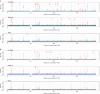
Cost-effective oligonucleotide genotyping arrays like the Affymetrix SNP 6.0 are still the predominant technique to measure DNA copy number variations (CNVs). However, CNV detection methods for microarrays overestimate both the number and the size of CNV regions and, consequently, suffer from a high false discovery rate (FDR). A high FDR means that many CNVs are wrongly detected and therefore not associated with a disease in a clinical study, though correction for multiple testing takes them into account and thereby decreases the study's discovery power. For controlling the FDR, we propose a probabilistic latent variable model, 'cn.FARMS', which is optimized by a Bayesian maximum a posteriori approach. cn.FARMS controls the FDR through the information gain of the posterior over the prior. The prior represents the null hypothesis of copy number 2 for all samples from which the posterior can only deviate by strong and consistent signals in the data. On HapMap data, cn.FARMS clearly outperformed the two most prevalent methods with respect to sensitivity and FDR. The software cn.FARMS is publicly available as a R package at http://www.bioinf.jku.at/software/cnfarms/cnfarms.html.
Figures




References
-
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung H-C, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. - PubMed
-
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

