Determinants of translation efficiency and accuracy
- PMID: 21487400
- PMCID: PMC3101949
- DOI: 10.1038/msb.2011.14
Determinants of translation efficiency and accuracy
Abstract
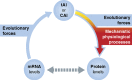
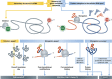
Proper functioning of biological cells requires that the process of protein expression be carried out with high efficiency and fidelity. Given an amino-acid sequence of a protein, multiple degrees of freedom still remain that may allow evolution to tune efficiency and fidelity for each gene under various conditions and cell types. Particularly, the redundancy of the genetic code allows the choice between alternative codons for the same amino acid, which, although 'synonymous,' may exert dramatic effects on the process of translation. Here we review modern developments in genomics and systems biology that have revolutionized our understanding of the multiple means by which translation is regulated. We suggest new means to model the process of translation in a richer framework that will incorporate information about gene sequences, the tRNA pool of the organism and the thermodynamic stability of the mRNA transcripts. A practical demonstration of a better understanding of the process would be a more accurate prediction of the proteome, given the transcriptome at a diversity of biological conditions.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

