Preparation and analysis of glycan microarrays
- PMID: 21488041
- PMCID: PMC3097418
- DOI: 10.1002/0471140864.ps1210s64
Preparation and analysis of glycan microarrays
Abstract
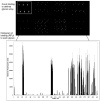
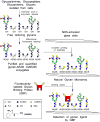
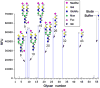
Determination of the binding specificity of glycan-binding proteins (GBPs), such as lectins, antibodies, and receptors, has traditionally been difficult and laborious. The advent of glycan microarrays has revolutionized the field of glycobiology by allowing simultaneous screening of a GBP for interactions with a large set of glycans in a single format. This unit describes the theory and method for production of two types of glycan microarrays (chemo/enzymatically synthesized and naturally derived), and their application to functional glycomics to explore glycan recognition by GBPs. These procedures are amenable to various types of arrays and a wide range of GBP samples.
© 2011 by John Wiley & Sons, Inc.
Figures

References
-
- Bohorov O, Andersson-Sand H, et al. Arraying glycomics: a novel bi-functional spacer for one-step microscale derivatization of free reducing glycans. Glycobiology. 2006;16:21C–27C. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

