Hendra and nipah infection: pathology, models and potential therapies
- PMID: 21488828
- PMCID: PMC3253017
- DOI: 10.2174/187152611795768097
Hendra and nipah infection: pathology, models and potential therapies
Abstract
The Paramyxoviridae family comprises of several genera that contain emerging or re-emerging threats for human and animal health with no real specific effective treatment available. Hendra and Nipah virus are members of a newly identified genus of emerging paramyxoviruses, Henipavirus. Since their discovery in the 1990s, henipaviruses outbreaks have been associated with high economic and public health threat potential. When compared to other paramyxoviruses, henipaviruses appear to have unique characteristics. Henipaviruses are zoonotic paramyxoviruses with a broader tropism than most other paramyxoviruses, and can cause severe acute encephalitis with unique features among viral encephalitides. There are currently no approved effective prophylactic or therapeutic treatments for henipavirus infections. Although ribavirin was empirically used and seemed beneficial during the biggest outbreak caused by one of these viruses, the Nipah virus, its efficacy is disputed in light of its lack of efficacy in several animal models of henipavirus infection. Nevertheless, because of its highly pathogenic nature, much effort has been spent in developing anti-henipavirus therapeutics. In this review we describe the unique features of henipavirus infections and the different strategies and animal models that have been developed so far in order to identify and test potential drugs to prevent or treat henipavirus infections. Some of these components have the potential to be broad-spectrum antivirals as they target effectors of viral pathogenecity common to other viruses. We will focus on small molecules or biologics, rather than vaccine strategies, that have been developed as anti-henipaviral therapeutics.
Figures
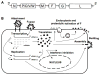
References
-
- CSIRO. Available from: http://www.csiro.au/science/Hendra-Virus.html.
-
- Lo MK, Rota PA. The emergence of Nipah virus, a highly pathogenic paramyxovirus. J Clin Virol. 2008;4:396–400. - PubMed
-
- Maisner A, Neufeld J, Weingartl H. Organ- and endotheliotropism of Nipah virus infections in vivo and in vitro. Thromb Haemost. 2009;6:1014–23. - PubMed
-
- Wild TF. Henipaviruses: a new family of emerging Paramyxoviruses. Pathol Biol (Paris) 2009;2:188–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

