Microbial shifts during dental biofilm re-development in the absence of oral hygiene in periodontal health and disease
- PMID: 21488936
- PMCID: PMC3177321
- DOI: 10.1111/j.1600-051X.2011.01730.x
Microbial shifts during dental biofilm re-development in the absence of oral hygiene in periodontal health and disease
Abstract
Aim: To monitor microbial shifts during dental biofilm re-development.
Materials and methods: Supra- and subgingival plaque samples were taken separately from 28 teeth in 38 healthy and 17 periodontitis subjects at baseline and immediately after tooth cleaning. Samples were taken again from seven teeth in randomly selected quadrants during 1, 2, 4 and 7 days of no oral hygiene. Samples were analysed using checkerboard DNA-DNA hybridization. Species counts were averaged within subjects at each time point. Significant differences in the counts between healthy and periodontitis subjects were determined using the Mann-Whitney test.
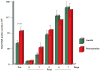
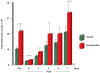
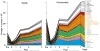
Results: The total supra- and subgingival counts were significantly higher in periodontitis on entry and reached or exceeded the baseline values after day 2. Supragingival counts of Veillonella parvula, Fusobacterium nucleatum ss vincentii and Neisseria mucosa increased from 2 to 7 days. Subgingival counts were greater for Actinomyces, green and orange complex species. Significant differences between groups in supragingival counts occurred for 17 of 41 species at entry, 0 at day 7; for subgingival plaque, these values were 39/41 taxa at entry, 17/41 at day 7.
Conclusions: Supragingival plaque re-development was similar in periodontitis and health, but subgingival species recolonization was more marked in periodontitis.
© 2011 John Wiley & Sons A/S.
Conflict of interest statement
Conflicts of Interest: The authors declare that they have no conflict of interests.
Figures



References
-
- Bernimoulin JP. Recent concepts in plaque formation. Journal of Clinical Periodontogy. 2003;30 5:7–9. - PubMed
-
- Gibbons RJ, Nygaard M. Interbacterial aggregation of plaque bacteria. Archives of Oral Biology. 1970;15:1397–1400. - PubMed
-
- Haffajee AD, Socransky SS, Goodson JM. Comparison of different data analyses for detecting changes in attachment level. Journal of Clinical Periodontology. 1983;10:298–310. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

