Genome-wide analysis of the 5' and 3' ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts
- PMID: 21490097
- PMCID: PMC3126295
- DOI: 10.1128/JVI.00428-11
Genome-wide analysis of the 5' and 3' ends of vaccinia virus early mRNAs delineates regulatory sequences of annotated and anomalous transcripts
Abstract
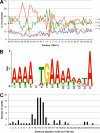
Poxviruses are large DNA viruses that encode a multisubunit RNA polymerase, stage-specific transcription factors, and enzymes that cap and polyadenylate mRNAs within the cytoplasm of infected animal cells. Genome-wide microarray and RNA-seq technologies have been used to profile the transcriptome of vaccinia virus (VACV), the prototype member of the family. Here, we adapted tag-based methods in conjunction with SOLiD and Illumina deep sequencing platforms to determine the precise 5' and 3' ends of VACV early mRNAs and map the putative transcription start sites (TSSs) and polyadenylation sites (PASs). Individual and clustered TSSs were found preceding 104 annotated open reading frames (ORFs), excluding pseudogenes. In the majority of cases, a 15-nucleotide consensus core motif was present upstream of the ORF. This motif, however, was also present at numerous other locations, indicating that it was insufficient for transcription initiation. Further analysis revealed a 10-nucleotide AT-rich spacer following functional core motifs that may facilitate DNA unwinding. Additional putative TSSs occurred in anomalous locations that may expand the functional repertoire of the VACV genome. However, many of the anomalous TSSs lacked an upstream core motif, raising the possibility that they arose by a processing mechanism as has been proposed for eukaryotic systems. Discrete and clustered PASs occurred about 40 nucleotides after an UUUUUNU termination signal. However, a large number of PASs were not preceded by this motif, suggesting alternative polyadenylation mechanisms. Pyrimidine-rich coding strand sequences were found immediately upstream of both types of PASs, signifying an additional feature of VACV 3'-end formation and polyadenylation.
Figures






References
-
- Ahn B.-Y., Gershon P. D., Moss B. 1994. RNA polymerase-associated protein RAP94 confers promoter specificity for initiating transcription of vaccinia virus early stage genes. J. Biol. Chem. 269:7552–7557 - PubMed
-
- Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 - PubMed
-
- Bailey T. L., Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

