Analysis of interferon signaling by infectious hepatitis C virus clones with substitutions of core amino acids 70 and 91
- PMID: 21490101
- PMCID: PMC3126319
- DOI: 10.1128/JVI.02583-10
Analysis of interferon signaling by infectious hepatitis C virus clones with substitutions of core amino acids 70 and 91
Abstract
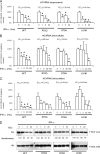
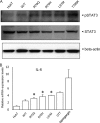
Substitution of amino acids 70 and 91 in the hepatitis C virus (HCV) core region is a significant predictor of poor responses to peginterferon-plus-ribavirin therapy, while their molecular mechanisms remain unclear. Here we investigated these differences in the response to alpha interferon (IFN) by using HCV cell culture with R70Q, R70H, and L91M substitutions. IFN treatment of cells transfected or infected with the wild type or the mutant HCV clones showed that the R70Q, R70H, and L91M core mutants were significantly more resistant than the wild type. Among HCV-transfected cells, intracellular HCV RNA levels were significantly higher for the core mutants than for the wild type, while HCV RNA in culture supernatant was significantly lower for these mutants than for the wild type. IFN-induced phosphorylation of STAT1 and STAT2 and expression of the interferon-inducible genes were significantly lower for the core mutants than for the wild type, suggesting cellular unresponsiveness to IFN. The expression level of an interferon signal attenuator, SOCS3, was significantly higher for the R70Q, R70H, and L91M mutants than for the wild type. Interleukin 6 (IL-6), which upregulates SOCS3, was significantly higher for the R70Q, R70H, and L91M mutants than for the wild type, suggesting interferon resistance, possibly through IL-6-induced, SOCS3-mediated suppression of interferon signaling. Expression levels of endoplasmic reticulum (ER) stress proteins were significantly higher in cells transfected with a core mutant than in those transfected with the wild type. In conclusion, HCV R70 and L91 core mutants were resistant to interferon in vitro, and the resistance may be induced by IL-6-induced upregulation of SOCS3. Those mechanisms may explain clinical interferon resistance of HCV core mutants.
Figures

Similar articles
-
Impact of amino acid substitutions in hepatitis C virus core region on the severe oxidative stress.Free Radic Biol Med. 2024 Feb 20;212:199-206. doi: 10.1016/j.freeradbiomed.2023.12.014. Epub 2023 Dec 14. Free Radic Biol Med. 2024. PMID: 38103659
-
Defective hepatic response to interferon and activation of suppressor of cytokine signaling 3 in chronic hepatitis C.Gastroenterology. 2007 Feb;132(2):733-44. doi: 10.1053/j.gastro.2006.11.045. Epub 2006 Nov 29. Gastroenterology. 2007. PMID: 17258724 Free PMC article.
-
Amino acid substitutions of hepatitis C virus core protein are not associated with intracellular antiviral response to interferon-α in vitro.Liver Int. 2010 Oct;30(9):1324-31. doi: 10.1111/j.1478-3231.2010.02299.x. Liver Int. 2010. PMID: 20602680
-
Before Direct-Acting Antivirals for Hepatitis C Virus: Evaluation of Core Protein R70Q and L/C91M Substitutions in Chronically Infected Brazilian Patients Unresponsive to IFN and/or RBV.Viruses. 2023 Jan 9;15(1):187. doi: 10.3390/v15010187. Viruses. 2023. PMID: 36680226 Free PMC article.
-
Understanding the molecular mechanism(s) of hepatitis C virus (HCV) induced interferon resistance.Infect Genet Evol. 2013 Oct;19:113-9. doi: 10.1016/j.meegid.2013.06.025. Epub 2013 Jul 5. Infect Genet Evol. 2013. PMID: 23831932 Review.
Cited by
-
Frequency of Interferon-Resistance Conferring Substitutions in Amino Acid Positions 70 and 91 of Core Protein of the Russian HCV 1b Isolates Analyzed in the T-Cell Epitopic Context.J Immunol Res. 2018 Feb 7;2018:7685371. doi: 10.1155/2018/7685371. eCollection 2018. J Immunol Res. 2018. PMID: 29577052 Free PMC article.
-
Chaperones in hepatitis C virus infection.World J Hepatol. 2016 Jan 8;8(1):9-35. doi: 10.4254/wjh.v8.i1.9. World J Hepatol. 2016. PMID: 26783419 Free PMC article. Review.
-
Substitution in Amino Acid 70 of Hepatitis C Virus Core Protein Changes the Adipokine Profile via Toll-Like Receptor 2/4 Signaling.PLoS One. 2015 Jun 29;10(6):e0131346. doi: 10.1371/journal.pone.0131346. eCollection 2015. PLoS One. 2015. PMID: 26121241 Free PMC article.
-
SOCS3 mRNA expression and polymorphisms as pretreatment predictor of response to HCV genotype 3a IFN-based treatment.Springerplus. 2016 Oct 21;5(1):1826. doi: 10.1186/s40064-016-3506-5. eCollection 2016. Springerplus. 2016. PMID: 27818864 Free PMC article.
-
Recent advances in understanding viral evasion of type I interferon.Immunology. 2013 Mar;138(3):190-7. doi: 10.1111/imm.12038. Immunology. 2013. PMID: 23173987 Free PMC article. Review.
References
-
- Akuta N., et al. 2010. Amino acid substitution in HCV core region and genetic variation near IL28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology 52:421–429 - PubMed
-
- Akuta N., et al. 2007. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology 46:1357–1364 - PubMed
-
- Akuta N., et al. 2008. Efficacy of low-dose intermittent interferon-alpha monotherapy in patients infected with hepatitis C virus genotype 1b who were predicted or failed to respond to pegylated interferon plus ribavirin combination therapy. J. Med. Virol. 80:1363–1369 - PubMed
-
- Akuta N., et al. 2007. Predictive factors of early and sustained responses to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b: amino acid substitutions in the core region and low-density lipoprotein cholesterol levels. J. Hepatol. 46:403–410 - PubMed
-
- Akuta N., et al. 2005. Virological and biochemical relapse after discontinuation of lamivudine monotherapy for chronic hepatitis B in Japan: comparison with breakthrough hepatitis during long-term treatment. Intervirology 48:174–182 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

