Modification site localization scoring integrated into a search engine
- PMID: 21490164
- PMCID: PMC3134073
- DOI: 10.1074/mcp.M111.008078
Modification site localization scoring integrated into a search engine
Abstract
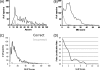
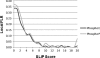
Large proteomic data sets identifying hundreds or thousands of modified peptides are becoming increasingly common in the literature. Several methods for assessing the reliability of peptide identifications both at the individual peptide or data set level have become established. However, tools for measuring the confidence of modification site assignments are sparse and are not often employed. A few tools for estimating phosphorylation site assignment reliabilities have been developed, but these are not integral to a search engine, so require a particular search engine output for a second step of processing. They may also require use of a particular fragmentation method and are mostly only applicable for phosphorylation analysis, rather than post-translational modifications analysis in general. In this study, we present the performance of site assignment scoring that is directly integrated into the search engine Protein Prospector, which allows site assignment reliability to be automatically reported for all modifications present in an identified peptide. It clearly indicates when a site assignment is ambiguous (and if so, between which residues), and reports an assignment score that can be translated into a reliability measure for individual site assignments.
Figures


References
-
- Witze E. S., Old W. M., Resing K. A., Ahn N. G. (2007) Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 4, 798–806 - PubMed
-
- Nilsson T., Mann M., Aebersold R., Yates J. R., 3rd, Bairoch A., Bergeron J. J. (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat. Methods 7, 681–685 - PubMed
-
- Nesvizhskii A. I., Vitek O., Aebersold R. (2007) Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

