Efficient silencing of gene expression by an ASON-bulge-DNAzyme complex
- PMID: 21490924
- PMCID: PMC3072403
- DOI: 10.1371/journal.pone.0018629
Efficient silencing of gene expression by an ASON-bulge-DNAzyme complex
Abstract
Background: DNAzymes are DNA molecules that can directly cleave cognate mRNA, and have been developed to silence gene expression for research and clinical purposes. The advantage of DNAzymes over ribozymes is that they are inexpensive to produce and exhibit good stability. The "10-23 DNA enzyme" is composed of a catalytic domain of 15 deoxynucleotides, flanked by two substrate-recognition domains of approximately eight nucleotides in each direction, which provides the complementary sequence required for specific binding to RNA substrates. However, these eight nucleotides might not afford sufficient binding energy to hold the RNA substrate along with the DNAzyme, which would interfere with the efficiency of the DNAzyme or cause side effects, such as the cleavage of non-cognate mRNAs.
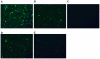
Methodology: In this study, we inserted a nonpairing bulge at the 5' end of the "10-23 DNA enzyme" to enhance its efficiency and specificity. Different sizes of bulges were inserted at different positions in the 5' end of the DNAzyme. The non-matching bulge will avoid strong binding between the DNAzyme and target mRNA, which may interfere with the efficiency of the DNAzyme.
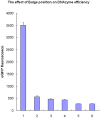
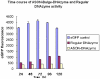
Conclusions: Our novel DNAzyme constructs could efficiently silence the expression of target genes, proving a powerful tool for gene silencing. The results showed that the six oligo bulge was the most effective when the six oligo bulge was 12-15 bp away from the core catalytic domain.
Conflict of interest statement
Figures



Similar articles
-
A complex RNA-cleaving DNAzyme that can efficiently cleave a pyrimidine-pyrimidine junction.J Mol Biol. 2010 Jul 23;400(4):689-701. doi: 10.1016/j.jmb.2010.05.047. Epub 2010 May 26. J Mol Biol. 2010. PMID: 20630470
-
Development of Visible-Light-Responsive RNA Scissors Based on a 10-23 DNAzyme.Chembiochem. 2018 Jun 18;19(12):1305-1311. doi: 10.1002/cbic.201800020. Epub 2018 May 30. Chembiochem. 2018. PMID: 29682882
-
Probing the function of nucleotides in the catalytic cores of the 8-17 and 10-23 DNAzymes by abasic nucleotide and C3 spacer substitutions.Biochemistry. 2010 Sep 7;49(35):7553-62. doi: 10.1021/bi100304b. Biochemistry. 2010. PMID: 20698496
-
Molecular Features and Metal Ions That Influence 10-23 DNAzyme Activity.Molecules. 2020 Jul 7;25(13):3100. doi: 10.3390/molecules25133100. Molecules. 2020. PMID: 32646019 Free PMC article. Review.
-
Theranostic DNAzymes.Theranostics. 2017 Feb 23;7(4):1010-1025. doi: 10.7150/thno.17736. eCollection 2017. Theranostics. 2017. PMID: 28382172 Free PMC article. Review.
Cited by
-
Studies on the Effect of Lipofectamine and Cell-Penetrating Peptide on the Properties of 10-23 DNAzyme.Molecules. 2023 May 7;28(9):3942. doi: 10.3390/molecules28093942. Molecules. 2023. PMID: 37175352 Free PMC article.
-
Emerging role of different DNA methyltransferases in the pathogenesis of cancer.Front Pharmacol. 2022 Aug 25;13:958146. doi: 10.3389/fphar.2022.958146. eCollection 2022. Front Pharmacol. 2022. PMID: 36091786 Free PMC article. Review.
References
-
- Bronson SK, Smithies O. Altering mice by homologous recombination using embryonic stem cells. J Biol Chem. 1994;269:27155–27158. - PubMed
-
- Izant JG, Weintraub H. Inhibition of thymidine kinase gene expression by anti-sense RNA: a molecular approach to genetic analysis. Cell. 1984;36:1007–1015. - PubMed
-
- Pieken WA, Olsen DB, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2′-modified hammerhead ribozymes. Science. 1991;253:314–317. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

