Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects
- PMID: 21490951
- PMCID: PMC3072368
- DOI: 10.1371/journal.pgen.1001362
Quantitative fitness analysis shows that NMD proteins and many other protein complexes suppress or enhance distinct telomere cap defects
Abstract
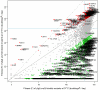
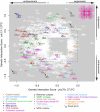
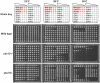
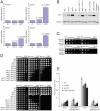
To better understand telomere biology in budding yeast, we have performed systematic suppressor/enhancer analyses on yeast strains containing a point mutation in the essential telomere capping gene CDC13 (cdc13-1) or containing a null mutation in the DNA damage response and telomere capping gene YKU70 (yku70Δ). We performed Quantitative Fitness Analysis (QFA) on thousands of yeast strains containing mutations affecting telomere-capping proteins in combination with a library of systematic gene deletion mutations. To perform QFA, we typically inoculate 384 separate cultures onto solid agar plates and monitor growth of each culture by photography over time. The data are fitted to a logistic population growth model; and growth parameters, such as maximum growth rate and maximum doubling potential, are deduced. QFA reveals that as many as 5% of systematic gene deletions, affecting numerous functional classes, strongly interact with telomere capping defects. We show that, while Cdc13 and Yku70 perform complementary roles in telomere capping, their genetic interaction profiles differ significantly. At least 19 different classes of functionally or physically related proteins can be identified as interacting with cdc13-1, yku70Δ, or both. Each specific genetic interaction informs the roles of individual gene products in telomere biology. One striking example is with genes of the nonsense-mediated RNA decay (NMD) pathway which, when disabled, suppress the conditional cdc13-1 mutation but enhance the null yku70Δ mutation. We show that the suppressing/enhancing role of the NMD pathway at uncapped telomeres is mediated through the levels of Stn1, an essential telomere capping protein, which interacts with Cdc13 and recruitment of telomerase to telomeres. We show that increased Stn1 levels affect growth of cells with telomere capping defects due to cdc13-1 and yku70Δ. QFA is a sensitive, high-throughput method that will also be useful to understand other aspects of microbial cell biology.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181–190. - PubMed
-
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. - PubMed
-
- Fisher TS, Zakian VA. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair (Amst) 2005;4:1215–1226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBF0235451/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 075294/WT_/Wellcome Trust/United Kingdom
- B/C008200/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F023545/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F006039/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

