Survival motor neuron protein regulates stem cell division, proliferation, and differentiation in Drosophila
- PMID: 21490958
- PMCID: PMC3072375
- DOI: 10.1371/journal.pgen.1002030
Survival motor neuron protein regulates stem cell division, proliferation, and differentiation in Drosophila
Abstract
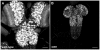
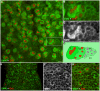
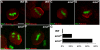
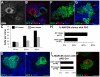
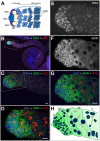
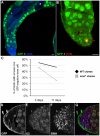
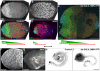
Spinal muscular atrophy is a severe neurogenic disease that is caused by mutations in the human survival motor neuron 1 (SMN1) gene. SMN protein is required for the assembly of small nuclear ribonucleoproteins and a dramatic reduction of the protein leads to cell death. It is currently unknown how the reduction of this ubiquitously essential protein can lead to tissue-specific abnormalities. In addition, it is still not known whether the disease is caused by developmental or degenerative defects. Using the Drosophila system, we show that SMN is enriched in postembryonic neuroblasts and forms a concentration gradient in the differentiating progeny. In addition to the developing Drosophila larval CNS, Drosophila larval and adult testes have a striking SMN gradient. When SMN is reduced in postembryonic neuroblasts using MARCM clonal analysis, cell proliferation and clone formation defects occur. These SMN mutant neuroblasts fail to correctly localise Miranda and have reduced levels of snRNAs. When SMN is removed, germline stem cells are lost more frequently. We also show that changes in SMN levels can disrupt the correct timing of cell differentiation. We conclude that highly regulated SMN levels are essential to drive timely cell proliferation and cell differentiation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat. 2000;15:228–237. - PubMed
-
- Brzustowicz LM, Lehner T, Castilla LH, Penchaszadeh GK, Wilhelmsen KC, et al. Genetic mapping of chronic childhood-onset spinal muscular atrophy to chromosome 5q11.2-13.3. Nature. 1990;344:540–541. - PubMed
-
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
-
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. - PubMed
-
- Lorson CL, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet. 2000;9:259–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

