Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome
- PMID: 21490963
- PMCID: PMC3072381
- DOI: 10.1371/journal.pgen.1002036
Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome
Abstract
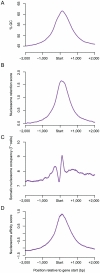
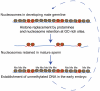
Chromatin in sperm is different from that in other cells, with most of the genome packaged by protamines not nucleosomes. Nucleosomes are, however, retained at some genomic sites, where they have the potential to transmit paternal epigenetic information. It is not understood how this retention is specified. Here we show that base composition is the major determinant of nucleosome retention in human sperm, predicting retention very well in both genic and non-genic regions of the genome. The retention of nucleosomes at GC-rich sequences with high intrinsic nucleosome affinity accounts for the previously reported retention at transcription start sites and at genes that regulate development. It also means that nucleosomes are retained at the start sites of most housekeeping genes. We also report a striking link between the retention of nucleosomes in sperm and the establishment of DNA methylation-free regions in the early embryo. Taken together, this suggests that paternal nucleosome transmission may facilitate robust gene regulation in the early embryo. We propose that chromatin organization in the male germline, rather than in somatic cells, is the major functional consequence of fine-scale base composition variation in the human genome. The selective pressure driving base composition evolution in mammals could, therefore, be the need to transmit paternal epigenetic information to the zygote.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Puwaravutipanich T, Panyim S. The nuclear basic proteins of human testes and ejaculated spermatozoa. Exp Cell Res. 1975;90:153–158. - PubMed
-
- Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–574. - PubMed
-
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 2010;17:679–687. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

