Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation
- PMID: 21490970
- PMCID: PMC3072388
- DOI: 10.1371/journal.pone.0018088
Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation
Abstract
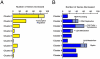
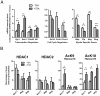
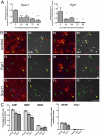
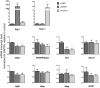
Differentiation of oligodendrocyte progenitor cells (OPCs) into mature oligodendrocytes requires extensive changes in gene expression, which are partly mediated by post-translational modifications of nucleosomal histones. An essential modification for oligodendrocyte differentiation is the removal of acetyl groups from lysine residues which is catalyzed by histone deacetylases (HDACs). The transcriptional targets of HDAC activity within OPCs however, have remained elusive and have been identified in this study by interrogating the oligodendrocyte transcriptome. Using a novel algorithm that allows clustering of gene transcripts according to expression kinetics and expression levels, we defined major waves of co-regulated genes. The initial overall decrease in gene expression was followed by the up-regulation of genes involved in lipid metabolism and myelination. Functional annotation of the down-regulated gene clusters identified transcripts involved in cell cycle regulation, transcription, and RNA processing. To define whether these genes were the targets of HDAC activity, we cultured rat OPCs in the presence of trichostatin A (TSA), an HDAC inhibitor previously shown to inhibit oligodendrocyte differentiation. By overlaying the defined oligodendrocyte transcriptome with the list of 'TSA sensitive' genes, we determined that a high percentage of 'TSA sensitive' genes are part of a normal program of oligodendrocyte differentiation. TSA treatment increased the expression of genes whose down-regulation occurs very early after induction of OPC differentiation, but did not affect the expression of genes with a slower kinetic. Among the increased 'TSA sensitive' genes we detected several transcription factors including Id2, Egr1, and Sox11, whose down-regulation is critical for OPC differentiation. Thus, HDAC target genes include clusters of co-regulated genes involved in transcriptional repression. These results support a de-repression model of oligodendrocyte lineage progression that relies on the concurrent down-regulation of several inhibitors of differentiation.
Conflict of interest statement
Figures


References
-
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. - PubMed
-
- Gielen E, Baron W, Vandeven M, Steels P, Hoekstra D, et al. Rafts in oligodendrocytes: evidence and structure-function relationship. Glia. 2006;54:499–512. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

