De novo polymerase activity and oligomerization of hepatitis C virus RNA-dependent RNA-polymerases from genotypes 1 to 5
- PMID: 21490973
- PMCID: PMC3072391
- DOI: 10.1371/journal.pone.0018515
De novo polymerase activity and oligomerization of hepatitis C virus RNA-dependent RNA-polymerases from genotypes 1 to 5
Abstract
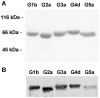
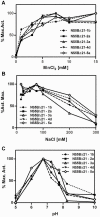
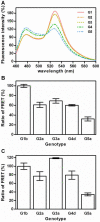
Hepatitis C virus (HCV) shows a great geographical diversity reflected in the high number of circulating genotypes and subtypes. The response to HCV treatment is genotype specific, with the predominant genotype 1 showing the lowest rate of sustained virological response. Virally encoded enzymes are candidate targets for intervention. In particular, promising antiviral molecules are being developed to target the viral NS3/4A protease and NS5B polymerase. Most of the studies with the NS5B polymerase have been done with genotypes 1b and 2a, whilst information about other genotypes is scarce. Here, we have characterized the de novo activity of NS5B from genotypes 1 to 5, with emphasis on conditions for optimum activity and kinetic constants. Polymerase cooperativity was determined by calculating the Hill coefficient and oligomerization through a new FRET-based method. The V(max)/K(m) ratios were statistically different between genotype 1 and the other genotypes (p<0.001), mainly due to differences in V(max) values, but differences in the Hill coefficient and NS5B oligomerization were noted. Analysis of sequence changes among the studied polymerases and crystal structures show the αF helix as a structural component probably involved in NS5B-NS5B interactions. The viability of the interaction of αF and αT helixes was confirmed by docking studies and calculation of electrostatic surface potentials for genotype 1 and point mutants corresponding to mutations from different genotypes. Results presented in this study reveal the existence of genotypic differences in NS5B de novo activity and oligomerization. Furthermore, these results allow us to define two regions, one consisting of residues Glu128, Asp129, and Glu248, and the other consisting of residues of αT helix possibly involved in NS5B-NS5B interactions.
Conflict of interest statement
Figures

References
-
- Perelson AS, Herrmann E, Micol F, Zeuzem S. New kinetic models for the hepatitis C virus. Hepatol. 2005;42:749–754. - PubMed
-
- Kuiken C, Yusim K, Boykin L, Richardson R. The Los Alamos HCV Sequence Database. Bioinformatics. 2005;21:379–384. - PubMed
-
- Kuiken C, Simmonds P. Nomenclature and numbering of the hepatitis C virus. Methods Mol Biol. 2009;510:33–53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

