The role of vimentin as a methylation biomarker for early diagnosis of cervical cancer
- PMID: 21491170
- PMCID: PMC3887602
- DOI: 10.1007/s10059-011-0229-x
The role of vimentin as a methylation biomarker for early diagnosis of cervical cancer
Abstract
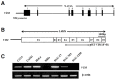
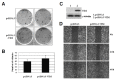
Multiple cytosine guanine dinucleotides (CpG island) are found in the VIM promoter region. The levels of VIM promoter methylation and VIM gene expression were investigated in 7 cervical cancer cell lines and 50 human tissue samples with a distinctive degree of malignant trans-formation. While multiple CpG sites in the VIM promoter were highly methylated in CIN III and invasive carcinoma cells, they were rarely methylated in normal cells. Our result shows that methylation in the VIM promoter appears to start from CIN I and CIN II, relatively early stages of multistep carcinogenesis. This epigenetic alteration in VIM promoter suggests the availability as a biomarker for the early diagnosis and prevention of cervical cancer. We also show that hypermethylation in the VIM promoter is responsible for transcriptional silencing of the VIM gene in cervical cancer cells. In addition, our result shows that exogenous overexpression of the VIM gene in SiHa cervical cancer cells slightly activated cell proliferation and migration as shown in soft agar colony formation and migration assays.
Figures



Similar articles
-
The role of hLHX6-HMR as a methylation biomarker for early diagnosis of cervical cancer.Oncol Rep. 2010 Jun;23(6):1675-82. doi: 10.3892/or_00000811. Oncol Rep. 2010. PMID: 20428825
-
The role of ADCYAP1, adenylate cyclase activating polypeptide 1, as a methylation biomarker for the early detection of cervical cancer.Oncol Rep. 2011 Jan;25(1):245-52. Oncol Rep. 2011. PMID: 21109983
-
Epigenetic regulation of the potential tumor suppressor gene, hLHX6.1, in human cervical cancer.Int J Oncol. 2011 Mar;38(3):859-69. doi: 10.3892/ijo.2011.904. Epub 2011 Jan 14. Int J Oncol. 2011. PMID: 21240459
-
TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia.J Natl Cancer Inst. 2004 Feb 18;96(4):294-305. doi: 10.1093/jnci/djh031. J Natl Cancer Inst. 2004. PMID: 14970278
-
The use of host cell DNA methylation analysis in the detection and management of women with advanced cervical intraepithelial neoplasia: a review.BJOG. 2021 Feb;128(3):504-514. doi: 10.1111/1471-0528.16395. Epub 2020 Aug 9. BJOG. 2021. PMID: 32619334 Free PMC article. Review.
Cited by
-
High Resolution Based Quantitative Determination of Methylation Status of CDH1 and VIM Gene in Epithelial Ovarian Cancer.Asian Pac J Cancer Prev. 2019 Oct 1;20(10):2923-2928. doi: 10.31557/APJCP.2019.20.10.2923. Asian Pac J Cancer Prev. 2019. PMID: 31653136 Free PMC article.
-
Aspects of the Epigenetic Regulation of EMT Related to Cancer Metastasis.Cells. 2021 Dec 6;10(12):3435. doi: 10.3390/cells10123435. Cells. 2021. PMID: 34943943 Free PMC article. Review.
-
Vimentin epigenetic deregulation in Bladder Cancer associates with acquisition of invasive and metastatic phenotype through epithelial-to-mesenchymal transition.Int J Biol Sci. 2023 Jan 1;19(1):1-12. doi: 10.7150/ijbs.77181. eCollection 2023. Int J Biol Sci. 2023. PMID: 36594099 Free PMC article.
-
Differential Expression of ADAM23, CDKN2A (P16), MMP14 and VIM Associated with Giant Cell Tumor of Bone.J Cancer. 2015 May 23;6(7):593-603. doi: 10.7150/jca.11238. eCollection 2015. J Cancer. 2015. PMID: 26078788 Free PMC article.
-
Single-cell transcriptome provides novel insights into antler stem cells, a cell type capable of mammalian organ regeneration.Funct Integr Genomics. 2019 Jul;19(4):555-564. doi: 10.1007/s10142-019-00659-2. Epub 2019 Jan 23. Funct Integr Genomics. 2019. PMID: 30673893
References
-
- Agorastos T., Miliaras D., Lambropoulos A.F., Chrisafi S., Kotsis A., Manthos A., Bontis J. Detection and typing of human papillomavirus DNA in uterine cervices with coexistent grade I and grade III intraepithelial neoplasia: biologic progression or independent lesions? Eur. J. Obstet. Gynecol. Reprod. (2005);121:99–103. - PubMed
-
- Baylin S.B., Ohm J.E. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. (2006);6:107–116. - PubMed
-
- Bosch F.X., Manos M.M., Munoz N., Sherman M., Jansen A.M., Peto J., Schiffman M.H., Moreno V., Kurman R., Shah K.V. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J. Natl. Cancer Inst. (1995);87:796–802. - PubMed
-
- Chen W.D., Han Z.J., Skoletsky J., Olson J., Sah J., Myeroff L. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J. Natl. Cancer Inst. (2005);97:1124–1132. - PubMed
-
- Costa V.L., Henrique R., Danielsen SA., Duarte-Pereira S., Eknaes M., Skotheim R.I., Rodrigues A., Magalhães J.S., Oliveira J., Lothe R.A., et al. Three epigenetic biomarkers, GDF15, TMEFF2 and VIM, accurately predict bladder cancer from DNA-based analyses of urine samples. Clin. Cancer Res. (2010);16:5842–5851. - PubMed

