Kinetic mechanism for the excision of hypoxanthine by Escherichia coli AlkA and evidence for binding to DNA ends
- PMID: 21491902
- PMCID: PMC3098123
- DOI: 10.1021/bi200232c
Kinetic mechanism for the excision of hypoxanthine by Escherichia coli AlkA and evidence for binding to DNA ends
Abstract
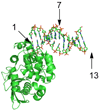
The Escherichia coli 3-methyladenine DNA glycosylase II protein (AlkA) recognizes a broad range of oxidized and alkylated base lesions and catalyzes the hydrolysis of the N-glycosidic bond to initiate the base excision repair pathway. Although the enzyme was one of the first DNA repair glycosylases to be discovered more than 25 years ago and there are multiple crystal structures, the mechanism is poorly understood. Therefore, we have characterized the kinetic mechanism for the AlkA-catalyzed excision of the deaminated purine, hypoxanthine. The multiple-turnover glycosylase assays are consistent with Michaelis-Menten kinetics. However, under single-turnover conditions that are commonly employed for studying other DNA glycosylases, we observe an unusual biphasic protein saturation curve. Initially, the observed rate constant for excision increases with an increasing level of AlkA protein, but at higher protein concentrations, the rate constant decreases. This behavior can be most easily explained by tight binding to DNA ends and by crowding of multiple AlkA protamers on the DNA. Consistent with this model, crystal structures have shown the preferential binding of AlkA to DNA ends. By varying the position of the lesion, we identified an asymmetric substrate that does not show inhibition at higher concentrations of AlkA, and we performed pre-steady state and steady state kinetic analysis. Unlike the situation in other glycosylases, release of the abasic product is faster than N-glycosidic bond cleavage. Nevertheless, AlkA exhibits significant product inhibition under multiple-turnover conditions, and it binds approximately 10-fold more tightly to an abasic site than to a hypoxanthine lesion site. This tight binding could help protect abasic sites when the adaptive response to DNA alkylation is activated and very high levels of AlkA protein are present.
Figures





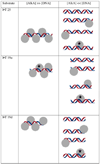
 ). If there is limiting amount of AlkA ([AlkA]<<[DNA]), then there will be an ensemble of DNA with AlkA bound at different binding sites across the DNA and every site can be sampled. For simplicity, only a few possible bound states are shown.
). If there is limiting amount of AlkA ([AlkA]<<[DNA]), then there will be an ensemble of DNA with AlkA bound at different binding sites across the DNA and every site can be sampled. For simplicity, only a few possible bound states are shown.

References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Gilchrest BA, Bohr VA. Aging processes, DNA damage, and repair. FASEB J. 1997;11:322–330. - PubMed
-
- Thomas L, Yang CH, Goldthwait DA. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982;21:1162–1169. - PubMed
-
- Lindahl T, Sedgwick B, Sekiguchi M, Nakabeppu Y. Regulation and expression of the adaptive response to alkylating agents. Annu. Rev. Biochem. 1988;57:133–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

