Transcriptional regulators of the GAD acid stress island are carried by effector protein-encoding prophages and indirectly control type III secretion in enterohemorrhagic Escherichia coli O157:H7
- PMID: 21492263
- PMCID: PMC7099609
- DOI: 10.1111/j.1365-2958.2011.07650.x
Transcriptional regulators of the GAD acid stress island are carried by effector protein-encoding prophages and indirectly control type III secretion in enterohemorrhagic Escherichia coli O157:H7
Abstract
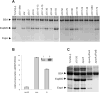
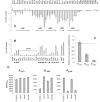
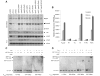
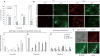
Type III secretion (T3S) plays a pivotal role in the colonization of ruminant hosts by Enterohemorrhagic Escherichia coli (EHEC). The T3S system translocates effector proteins into host cells to promote bacterial attachment and persistence. The repertoire and variation in prophage regions underpins differences in the pathogenesis and epidemiology of EHEC strains. In this study, we have used a collection of deletions in cryptic prophages and EHEC O157 O-islands to screen for novel regulators of T3S. Using this approach we have identified a family of homologous AraC-like regulators that indirectly repress T3S. These prophage-encoded secretion regulator genes (psr) are found exclusively on prophages and are associated with effector loci and the T3S activating Pch family of regulators. Transcriptional profiling, mutagenesis and DNA binding studies were used to show that these regulators usurp the conserved GAD acid stress resistance system to regulate T3S by increasing the expression of GadE (YhiE) and YhiF and that this regulation follows attachment to bovine epithelial cells. We further demonstrate that PsrA and effectors encoded within cryptic prophage CP933-N are required for persistence in a ruminant model of colonization.
© 2011 Blackwell Publishing Ltd.
Figures



Similar articles
-
Identification of a novel prophage regulator in Escherichia coli controlling the expression of type III secretion.Mol Microbiol. 2012 Jan;83(1):208-23. doi: 10.1111/j.1365-2958.2011.07927.x. Epub 2011 Dec 9. Mol Microbiol. 2012. PMID: 22111928 Free PMC article.
-
Control of acid resistance pathways of enterohemorrhagic Escherichia coli strain EDL933 by PsrB, a prophage-encoded AraC-like regulator.Infect Immun. 2015 Jan;83(1):346-53. doi: 10.1128/IAI.02758-14. Epub 2014 Nov 3. Infect Immun. 2015. PMID: 25368119 Free PMC article.
-
Sequence variability of P2-like prophage genomes carrying the cytolethal distending toxin V operon in Escherichia coli O157.Appl Environ Microbiol. 2013 Aug;79(16):4958-64. doi: 10.1128/AEM.01134-13. Epub 2013 Jun 14. Appl Environ Microbiol. 2013. PMID: 23770900 Free PMC article.
-
Controlling injection: regulation of type III secretion in enterohaemorrhagic Escherichia coli.Trends Microbiol. 2009 Aug;17(8):361-70. doi: 10.1016/j.tim.2009.06.001. Epub 2009 Aug 5. Trends Microbiol. 2009. PMID: 19660954 Review.
-
SdiA sensing of acyl-homoserine lactones by enterohemorrhagic E. coli (EHEC) serotype O157:H7 in the bovine rumen.Gut Microbes. 2010 Nov-Dec;1(6):432-5. doi: 10.4161/gmic.1.6.14177. Gut Microbes. 2010. PMID: 21468228 Free PMC article. Review.
Cited by
-
Identification of a novel prophage regulator in Escherichia coli controlling the expression of type III secretion.Mol Microbiol. 2012 Jan;83(1):208-23. doi: 10.1111/j.1365-2958.2011.07927.x. Epub 2011 Dec 9. Mol Microbiol. 2012. PMID: 22111928 Free PMC article.
-
Control of acid resistance pathways of enterohemorrhagic Escherichia coli strain EDL933 by PsrB, a prophage-encoded AraC-like regulator.Infect Immun. 2015 Jan;83(1):346-53. doi: 10.1128/IAI.02758-14. Epub 2014 Nov 3. Infect Immun. 2015. PMID: 25368119 Free PMC article.
-
Virulence-related O islands in enterohemorrhagic Escherichia coli O157:H7.Gut Microbes. 2021 Jan-Dec;13(1):1992237. doi: 10.1080/19490976.2021.1992237. Gut Microbes. 2021. PMID: 34711138 Free PMC article.
-
An O island 172 encoded RNA helicase regulates the motility of Escherichia coli O157:H7.PLoS One. 2013 Jun 13;8(6):e64211. doi: 10.1371/journal.pone.0064211. Print 2013. PLoS One. 2013. PMID: 23785398 Free PMC article.
-
Regulation of the Locus of Enterocyte Effacement in Attaching and Effacing Pathogens.J Bacteriol. 2017 Dec 20;200(2):e00336-17. doi: 10.1128/JB.00336-17. Print 2018 Jan 15. J Bacteriol. 2017. PMID: 28760850 Free PMC article. Review.
References
-
- Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JA, Molin S, Prensier G, Arbeille B, Ghigo JM. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol. 2004;51:659–674. - PubMed
-
- Bustamante VH, Santana FJ, Calva E, Puente JL. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol. 2001;39:664–678. - PubMed
-
- Campellone KG, Robbins D, Leong JM. EspF(U) is a translocated EHEC effector that interacts with Tir and N-WASP and promotes nck-independent actin assembly. Developmental Cell. 2004;7:217–228. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

