A 6-gene signature identifies four molecular subgroups of neuroblastoma
- PMID: 21492432
- PMCID: PMC3095533
- DOI: 10.1186/1475-2867-11-9
A 6-gene signature identifies four molecular subgroups of neuroblastoma
Abstract
Background: There are currently three postulated genomic subtypes of the childhood tumour neuroblastoma (NB); Type 1, Type 2A, and Type 2B. The most aggressive forms of NB are characterized by amplification of the oncogene MYCN (MNA) and low expression of the favourable marker NTRK1. Recently, mutations or high expression of the familial predisposition gene Anaplastic Lymphoma Kinase (ALK) was associated to unfavourable biology of sporadic NB. Also, various other genes have been linked to NB pathogenesis.
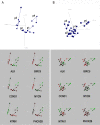
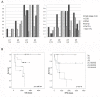
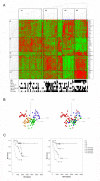
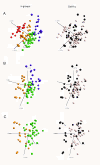
Results: The present study explores subgroup discrimination by gene expression profiling using three published microarray studies on NB (47 samples). Four distinct clusters were identified by Principal Components Analysis (PCA) in two separate data sets, which could be verified by an unsupervised hierarchical clustering in a third independent data set (101 NB samples) using a set of 74 discriminative genes. The expression signature of six NB-associated genes ALK, BIRC5, CCND1, MYCN, NTRK1, and PHOX2B, significantly discriminated the four clusters (p < 0.05, one-way ANOVA test). PCA clusters p1, p2, and p3 were found to correspond well to the postulated subtypes 1, 2A, and 2B, respectively. Remarkably, a fourth novel cluster was detected in all three independent data sets. This cluster comprised mainly 11q-deleted MNA-negative tumours with low expression of ALK, BIRC5, and PHOX2B, and was significantly associated with higher tumour stage, poor outcome and poor survival compared to the Type 1-corresponding favourable group (INSS stage 4 and/or dead of disease, p < 0.05, Fisher's exact test).
Conclusions: Based on expression profiling we have identified four molecular subgroups of neuroblastoma, which can be distinguished by a 6-gene signature. The fourth subgroup has not been described elsewhere, and efforts are currently made to further investigate this group's specific characteristics.
Figures

References
-
- Brodeur GM, Pritchard J, Berthold F, Carlsen NL, Castel V, Castelberry RP, De Bernardi B, Evans AE, Favrot M, Hedborg F. et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–1477. - PubMed
-
- Breslow N, McCann B. Statistical estimation of prognosis for children with neuroblastoma. Cancer Res. 1971;31:2098–2103. - PubMed
-
- Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B, Stram DO, Gerbing RB, Lukens JN, Matthay KK, Castleberry RP. The International Neuroblastoma Pathology Classification (the Shimada system) Cancer. 1999;86:364–372. doi: 10.1002/(SICI)1097-0142(19990715)86:2<364::AID-CNCR21>3.0.CO;2-7. - DOI - PubMed
-
- Look AT, Hayes FA, Shuster JJ, Douglass EC, Castleberry RP, Bowman LC, Smith EI, Brodeur GM. Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastoma: a Pediatric Oncology Group study. J Clin Oncol. 1991;9:581–591. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

