Genomic insight into the common carp (Cyprinus carpio) genome by sequencing analysis of BAC-end sequences
- PMID: 21492448
- PMCID: PMC3083359
- DOI: 10.1186/1471-2164-12-188
Genomic insight into the common carp (Cyprinus carpio) genome by sequencing analysis of BAC-end sequences
Abstract
Background: Common carp is one of the most important aquaculture teleost fish in the world. Common carp and other closely related Cyprinidae species provide over 30% aquaculture production in the world. However, common carp genomic resources are still relatively underdeveloped. BAC end sequences (BES) are important resources for genome research on BAC-anchored genetic marker development, linkage map and physical map integration, and whole genome sequence assembling and scaffolding.
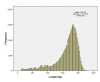
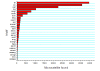
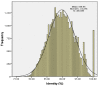
Result: To develop such valuable resources in common carp (Cyprinus carpio), a total of 40,224 BAC clones were sequenced on both ends, generating 65,720 clean BES with an average read length of 647 bp after sequence processing, representing 42,522,168 bp or 2.5% of common carp genome. The first survey of common carp genome was conducted with various bioinformatics tools. The common carp genome contains over 17.3% of repetitive elements with GC content of 36.8% and 518 transposon ORFs. To identify and develop BAC-anchored microsatellite markers, a total of 13,581 microsatellites were detected from 10,355 BES. The coding region of 7,127 genes were recognized from 9,443 BES on 7,453 BACs, with 1,990 BACs have genes on both ends. To evaluate the similarity to the genome of closely related zebrafish, BES of common carp were aligned against zebrafish genome. A total of 39,335 BES of common carp have conserved homologs on zebrafish genome which demonstrated the high similarity between zebrafish and common carp genomes, indicating the feasibility of comparative mapping between zebrafish and common carp once we have physical map of common carp.
Conclusion: BAC end sequences are great resources for the first genome wide survey of common carp. The repetitive DNA was estimated to be approximate 28% of common carp genome, indicating the higher complexity of the genome. Comparative analysis had mapped around 40,000 BES to zebrafish genome and established over 3,100 microsyntenies, covering over 50% of the zebrafish genome. BES of common carp are tremendous tools for comparative mapping between the two closely related species, zebrafish and common carp, which should facilitate both structural and functional genome analysis in common carp.
Figures
References
-
- Danzmann R, Davidson E, Ferguson M, Gharbi K, Koop B, Hoyheim B, Lien S, Lubieniecki K, Moghadam H, Park J. et al.Distribution of ancestral proto-Actinopterygian chromosome arms within the genomes of 4R-derivative salmonid fishes (Rainbow trout and Atlantic salmon) BMC Genomics. 2008;9(1):557. doi: 10.1186/1471-2164-9-557. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

