Centriole assembly and the role of Mps1: defensible or dispensable?
- PMID: 21492451
- PMCID: PMC3094359
- DOI: 10.1186/1747-1028-6-9
Centriole assembly and the role of Mps1: defensible or dispensable?
Abstract
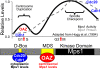
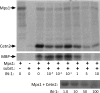
The Mps1 protein kinase is an intriguing and controversial player in centriole assembly. Originally shown to control duplication of the budding yeast spindle pole body, Mps1 is present in eukaryotes from yeast to humans, the nematode C. elegans being a notable exception, and has also been shown to regulate the spindle checkpoint and an increasing number of cellular functions relating to genomic stability. While its function in the spindle checkpoint appears to be both universally conserved and essential in most organisms, conservation of its originally described function in spindle pole duplication has proven controversial, and it is less clear whether Mps1 is essential for centrosome duplication outside of budding yeast. Recent studies of Mps1 have identified at least two distinct functions for Mps1 in centriole assembly, while simultaneously supporting the notion that Mps1 is dispensable for the process. However, the fact that at least one centrosomal substrate of Mps1 is conserved from yeast to humans down to the phosphorylation site, combined with evidence demonstrating the exquisite control exerted over centrosomal Mps1 levels suggest that the notion of being essential may not be the most important of distinctions.
Figures
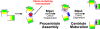

Similar articles
-
The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p.J Cell Biol. 2002 Feb 4;156(3):453-65. doi: 10.1083/jcb.200111025. Epub 2002 Feb 4. J Cell Biol. 2002. PMID: 11827982 Free PMC article.
-
The mitotic arrest in response to hypoxia and of polar bodies during early embryogenesis requires Drosophila Mps1.Curr Biol. 2004 Nov 23;14(22):2019-24. doi: 10.1016/j.cub.2004.11.008. Curr Biol. 2004. PMID: 15556864
-
Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly.Mol Biol Cell. 2010 Dec;21(24):4361-72. doi: 10.1091/mbc.E10-04-0298. Epub 2010 Oct 27. Mol Biol Cell. 2010. PMID: 20980622 Free PMC article.
-
Killing two birds with one stone: how budding yeast Mps1 controls chromosome segregation and spindle assembly checkpoint through phosphorylation of a single kinetochore protein.Curr Genet. 2020 Dec;66(6):1037-1044. doi: 10.1007/s00294-020-01091-x. Epub 2020 Jul 6. Curr Genet. 2020. PMID: 32632756 Review.
-
Spindle regulation: Mps1 flies into new areas.Curr Biol. 2004 Dec 29;14(24):R1058-60. doi: 10.1016/j.cub.2004.11.047. Curr Biol. 2004. PMID: 15620641 Review.
Cited by
-
Role of E2Fs and mitotic regulators controlled by E2Fs in the epithelial to mesenchymal transition.Exp Biol Med (Maywood). 2019 Nov;244(16):1419-1429. doi: 10.1177/1535370219881360. Epub 2019 Oct 1. Exp Biol Med (Maywood). 2019. PMID: 31575294 Free PMC article. Review.
-
A crystal structure of the human protein kinase Mps1 reveals an ordered conformation of the activation loop.Proteins. 2019 Apr;87(4):348-352. doi: 10.1002/prot.25651. Epub 2019 Jan 8. Proteins. 2019. PMID: 30582207 Free PMC article.
-
The MPS1 family of protein kinases.Annu Rev Biochem. 2012;81:561-85. doi: 10.1146/annurev-biochem-061611-090435. Epub 2012 Apr 5. Annu Rev Biochem. 2012. PMID: 22482908 Free PMC article. Review.
-
Crosstalk between cilia and autophagy: implication for human diseases.Autophagy. 2023 Jan;19(1):24-43. doi: 10.1080/15548627.2022.2067383. Epub 2022 May 25. Autophagy. 2023. PMID: 35613303 Free PMC article. Review.
-
Protein kinase TTK promotes proliferation and migration and mediates epithelial-mesenchymal transition in human bladder cancer cells.Int J Clin Exp Pathol. 2018 Oct 1;11(10):4854-4861. eCollection 2018. Int J Clin Exp Pathol. 2018. PMID: 31949560 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

