Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression
- PMID: 21492895
- PMCID: PMC3101335
- DOI: 10.1016/j.virol.2011.03.019
Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression
Abstract
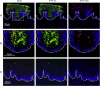
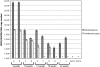
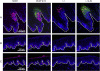
Rabbit oral papillomavirus (ROPV) causes benign and spontaneously regressing oral lesions in rabbits, and is a useful model of disease associated with low-risk human papillomavirus types. Here we have adapted the ROPV system to study papillomavirus latency. Following lesion regression, ROPV DNA persists at the majority of regressed sites at levels substantially lower than those found in productive papillomas. Spliced viral transcripts were also detected. ROPV persistence in the absence of disease could be demonstrated for a year following infection and lesion-regression. This was not associated with completion of the virus life-cycle or new virion production, indicating that ROPV persists in a latent state. Using novel laser capture microdissection techniques, we could show that the site of latency is a subset of basal epithelial cells at sites of previous experimental infection. We hypothesize that these cells are epithelial stem cells and that reactivation of latency may be a source of recurrent disease.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



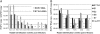

References
-
- Abramson A.L., Nouri M., Mullooly V., Fisch G., Steinberg B.M. Latent human papillomavirus infection is comparable in the larynx and trachea. J. Med. Virol. 2004;72(3):473–477. - PubMed
-
- Broker T.R., Jin, Croom-Rivers A., Bragg S.M., Richardson M., Chow L.T., Vermund S.H., Alvarez R.D., Pappas P.G., Squires K.E., Hoesley C.J. Viral latency—the papillomavirus model. Dev Biol (Basel) 2001;106:443–451. discussion 452–3, 465–75. - PubMed
-
- Christensen N.D., Cladel N.M., Reed C.A., Han R. Rabbit oral papillomavirus complete genome sequence and immunity following genital infection. Virology. 2000;269(2):451–461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

