Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting
- PMID: 21493687
- PMCID: PMC3130291
- DOI: 10.1093/nar/gkr218
Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting
Erratum in
- Nucleic Acids Res. 2011 Sep 1;39(17):7879
Abstract
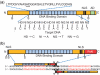
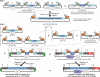
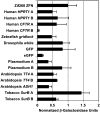
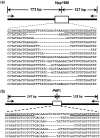
TALENs are important new tools for genome engineering. Fusions of transcription activator-like (TAL) effectors of plant pathogenic Xanthomonas spp. to the FokI nuclease, TALENs bind and cleave DNA in pairs. Binding specificity is determined by customizable arrays of polymorphic amino acid repeats in the TAL effectors. We present a method and reagents for efficiently assembling TALEN constructs with custom repeat arrays. We also describe design guidelines based on naturally occurring TAL effectors and their binding sites. Using software that applies these guidelines, in nine genes from plants, animals and protists, we found candidate cleavage sites on average every 35 bp. Each of 15 sites selected from this set was cleaved in a yeast-based assay with TALEN pairs constructed with our reagents. We used two of the TALEN pairs to mutate HPRT1 in human cells and ADH1 in Arabidopsis thaliana protoplasts. Our reagents include a plasmid construct for making custom TAL effectors and one for TAL effector fusions to additional proteins of interest. Using the former, we constructed de novo a functional analog of AvrHah1 of Xanthomonas gardneri. The complete plasmid set is available through the non-profit repository AddGene and a web-based version of our software is freely accessible online.
Figures


References
-
- Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 2010;13:394–401. - PubMed
-
- Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48:419–436. - PubMed
-
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. - PubMed
-
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

