Determination of structural models of the complex between the cytoplasmic domain of erythrocyte band 3 and ankyrin-R repeats 13-24
- PMID: 21493712
- PMCID: PMC3121456
- DOI: 10.1074/jbc.M111.230326
Determination of structural models of the complex between the cytoplasmic domain of erythrocyte band 3 and ankyrin-R repeats 13-24
Abstract
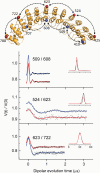
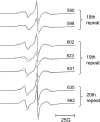
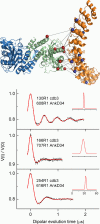
The adaptor protein ankyrin-R interacts via its membrane binding domain with the cytoplasmic domain of the anion exchange protein (AE1) and via its spectrin binding domain with the spectrin-based membrane skeleton in human erythrocytes. This set of interactions provides a bridge between the lipid bilayer and the membrane skeleton, thereby stabilizing the membrane. Crystal structures for the dimeric cytoplasmic domain of AE1 (cdb3) and for a 12-ankyrin repeat segment (repeats 13-24) from the membrane binding domain of ankyrin-R (AnkD34) have been reported. However, structural data on how these proteins assemble to form a stable complex have not been reported. In the current studies, site-directed spin labeling, in combination with electron paramagnetic resonance (EPR) and double electron-electron resonance, has been utilized to map the binding interfaces of the two proteins in the complex and to obtain inter-protein distance constraints. These data have been utilized to construct a family of structural models that are consistent with the full range of experimental data. These models indicate that an extensive area on the peripheral domain of cdb3 binds to ankyrin repeats 18-20 on the top loop surface of AnkD34 primarily through hydrophobic interactions. This is a previously uncharacterized surface for binding of cdb3 to AnkD34. Because a second dimer of cdb3 is known to bind to ankyrin repeats 7-12 of the membrane binding domain of ankyrin-R, the current models have significant implications regarding the structural nature of a tetrameric form of AE1 that is hypothesized to be involved in binding to full-length ankyrin-R in the erythrocyte membrane.
Figures
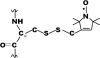







Similar articles
-
Identification of contact sites between ankyrin and band 3 in the human erythrocyte membrane.Biochemistry. 2012 Aug 28;51(34):6838-46. doi: 10.1021/bi300693k. Epub 2012 Aug 14. Biochemistry. 2012. PMID: 22861190 Free PMC article.
-
Identification of a critical ankyrin-binding loop on the cytoplasmic domain of erythrocyte membrane band 3 by crystal structure analysis and site-directed mutagenesis.J Biol Chem. 2003 Feb 28;278(9):6879-84. doi: 10.1074/jbc.M211137200. Epub 2002 Dec 12. J Biol Chem. 2003. PMID: 12482869
-
Rate of rupture and reattachment of the band 3-ankyrin bridge on the human erythrocyte membrane.J Biol Chem. 2006 Aug 4;281(31):22360-22366. doi: 10.1074/jbc.M513839200. Epub 2006 Jun 8. J Biol Chem. 2006. PMID: 16762928
-
The molecular basis for membrane - cytoskeleton association in human erythrocytes.J Cell Biochem. 1982;18(1):49-65. doi: 10.1002/jcb.1982.240180106. J Cell Biochem. 1982. PMID: 6461664 Review.
-
[Molecular defects of erythrocyte membrane proteins].Vopr Med Khim. 1996 Apr-Jun;42(2):103-10. Vopr Med Khim. 1996. PMID: 9148591 Review. Russian. No abstract available.
Cited by
-
Evidence of a structural and functional ammonium transporter RhBG·anion exchanger 1·ankyrin-G complex in kidney epithelial cells.J Biol Chem. 2015 Mar 13;290(11):6925-36. doi: 10.1074/jbc.M114.610048. Epub 2015 Jan 23. J Biol Chem. 2015. PMID: 25616663 Free PMC article.
-
Automated structure refinement for a protein heterodimer complex using limited EPR spectroscopic data and a rigid-body docking algorithm: a three-dimensional model for an ankyrin-CDB3 complex.J Phys Chem B. 2014 May 8;118(18):4717-26. doi: 10.1021/jp4099705. Epub 2014 Apr 23. J Phys Chem B. 2014. PMID: 24758720 Free PMC article.
-
A novel approach to modeling side chain ensembles of the bifunctional spin label RX.bioRxiv [Preprint]. 2023 May 24:2023.05.24.542139. doi: 10.1101/2023.05.24.542139. bioRxiv. 2023. PMID: 37292623 Free PMC article. Preprint.
-
Use of electron paramagnetic resonance to solve biochemical problems.Biochemistry. 2013 Sep 3;52(35):5967-84. doi: 10.1021/bi400834a. Epub 2013 Aug 20. Biochemistry. 2013. PMID: 23961941 Free PMC article. Review.
-
MtsslWizard: In Silico Spin-Labeling and Generation of Distance Distributions in PyMOL.Appl Magn Reson. 2012 Apr;42(3):377-391. doi: 10.1007/s00723-012-0314-0. Epub 2012 Feb 3. Appl Magn Reson. 2012. PMID: 22448103 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

