Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2
- PMID: 21493713
- PMCID: PMC3103319
- DOI: 10.1074/jbc.M110.205211
Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2
Abstract
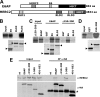
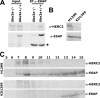
Deregulation of the ubiquitin-protein ligase E6AP contributes to the development of the Angelman syndrome and to cervical carcinogenesis suggesting that the activity of E6AP needs to be under tight control. However, how E6AP activity is regulated at the post-translational level under non-pathologic conditions is poorly understood. In this study, we report that the giant protein HERC2, which is like E6AP a member of the HECT family of ubiquitin-protein ligases, binds to E6AP. The interaction is mediated by the RCC1-like domain 2 of HERC2 and a region spanning amino acid residues 150-200 of E6AP. Furthermore, we provide evidence that HERC2 stimulates the ubiquitin-protein ligase activity of E6AP in vitro and within cells and that this stimulatory effect does not depend on the ubiquitin-protein ligase activity of HERC2. Thus, the data obtained indicate that HERC2 acts as a regulator of E6AP.
Figures

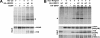
Similar articles
-
Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes.Mol Cell Biol. 2012 Aug;32(15):3095-106. doi: 10.1128/MCB.00201-12. Epub 2012 May 29. Mol Cell Biol. 2012. PMID: 22645313 Free PMC article.
-
The disordered negatively charged C-terminus of the large HECT E3 ubiquitin ligase HERC2 provides structural and thermal stability to the HECT C-lobe.Protein Sci. 2024 Dec;33(12):e5229. doi: 10.1002/pro.5229. Protein Sci. 2024. PMID: 39565083 Free PMC article.
-
The HERC2 ubiquitin ligase is essential for embryonic development and regulates motor coordination.Oncotarget. 2016 Aug 30;7(35):56083-56106. doi: 10.18632/oncotarget.11270. Oncotarget. 2016. PMID: 27528230 Free PMC article.
-
Physiological functions of the HECT family of ubiquitin ligases.Nat Rev Mol Cell Biol. 2009 Jun;10(6):398-409. doi: 10.1038/nrm2690. Epub 2009 May 13. Nat Rev Mol Cell Biol. 2009. PMID: 19436320 Review.
-
HECT E3 ubiquitin ligases - emerging insights into their biological roles and disease relevance.J Cell Sci. 2020 Apr 7;133(7):jcs228072. doi: 10.1242/jcs.228072. J Cell Sci. 2020. PMID: 32265230 Free PMC article. Review.
Cited by
-
The HECT E3 Ligase E6AP/UBE3A as a Therapeutic Target in Cancer and Neurological Disorders.Cancers (Basel). 2020 Jul 29;12(8):2108. doi: 10.3390/cancers12082108. Cancers (Basel). 2020. PMID: 32751183 Free PMC article. Review.
-
UBE3A reinstatement as a disease-modifying therapy for Angelman syndrome.Dev Med Child Neurol. 2021 Jul;63(7):802-807. doi: 10.1111/dmcn.14831. Epub 2021 Feb 4. Dev Med Child Neurol. 2021. PMID: 33543479 Free PMC article. Review.
-
Exploring the Roles of HERC2 and the NEDD4L HECT E3 Ubiquitin Ligase Subfamily in p53 Signaling and the DNA Damage Response.Front Oncol. 2021 Mar 31;11:659049. doi: 10.3389/fonc.2021.659049. eCollection 2021. Front Oncol. 2021. PMID: 33869064 Free PMC article. Review.
-
Antisense oligonucleotide therapy rescues disturbed brain rhythms and sleep in juvenile and adult mouse models of Angelman syndrome.Elife. 2023 Jan 3;12:e81892. doi: 10.7554/eLife.81892. Elife. 2023. PMID: 36594817 Free PMC article.
-
Neurodevelopmental disease: A molecular tightrope.Nature. 2015 Oct 1;526(7571):50-1. doi: 10.1038/526050b. Nature. 2015. PMID: 26432241 No abstract available.
References
-
- Kerscher O., Felberbaum R., Hochstrasser M. (2006) Annu. Rev. Cell Dev. Biol. 22, 159–180 - PubMed
-
- Deshaies R. J., Joazeiro C. A. (2009) Annu. Rev. Biochem. 78, 399–434 - PubMed
-
- Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell. Biol. 10, 398–409 - PubMed
-
- Matsuura T., Sutcliffe J. S., Fang P., Galjaard R. J., Jiang Y. H., Benton C. S., Rommens J. M., Beaudet A. L. (1997) Nat. Genet. 15, 74–77 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

