Peracetylated 4-fluoro-glucosamine reduces the content and repertoire of N- and O-glycans without direct incorporation
- PMID: 21493714
- PMCID: PMC3122228
- DOI: 10.1074/jbc.M110.194597
Peracetylated 4-fluoro-glucosamine reduces the content and repertoire of N- and O-glycans without direct incorporation
Abstract
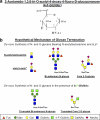
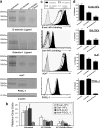
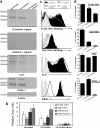
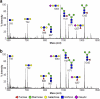
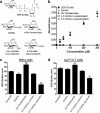
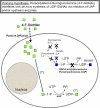
Prior studies have shown that treatment with the peracetylated 4-fluorinated analog of glucosamine (4-F-GlcNAc) elicits anti-skin inflammatory activity by ablating N-acetyllactosamine (LacNAc), sialyl Lewis X (sLe(X)), and related lectin ligands on effector leukocytes. Based on anti-sLe(X) antibody and lectin probing experiments on 4-F-GlcNAc-treated leukocytes, it was hypothesized that 4-F-GlcNAc inhibited sLe(X) formation by incorporating into LacNAc and blocking the addition of galactose or fucose at the carbon 4-position of 4-F-GlcNAc. To test this hypothesis, we determined whether 4-F-GlcNAc is directly incorporated into N- and O-glycans released from 4-F-GlcNAc-treated human sLe(X) (+) T cells and leukemic KG1a cells. At concentrations that abrogated galectin-1 (Gal-1) ligand and E-selectin ligand expression and related LacNAc and sLe(X) structures, MALDI-TOF and MALDI-TOF/TOF mass spectrometry analyses showed that 4-F-GlcNAc 1) reduced content and structural diversity of tri- and tetra-antennary N-glycans and of O-glycans, 2) increased biantennary N-glycans, and 3) reduced LacNAc and sLe(X) on N-glycans and on core 2 O-glycans. Moreover, MALDI-TOF MS did not reveal any m/z ratios relating to the presence of fluorine atoms, indicating that 4-F-GlcNAc did not incorporate into glycans. Further analysis showed that 4-F-GlcNAc treatment had minimal effect on expression of 1200 glycome-related genes and did not alter the activity of LacNAc-synthesizing enzymes. However, 4-F-GlcNAc dramatically reduced intracellular levels of uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc), a key precursor of LacNAc synthesis. These data show that Gal-1 and E-selectin ligand reduction by 4-F-GlcNAc is not caused by direct 4-F-GlcNAc glycan incorporation and consequent chain termination but rather by interference with UDP-GlcNAc synthesis.
Figures



References
-
- Liu F. T., Rabinovich G. A. (2010) Ann. N.Y. Acad. Sci. 1183, 158–182 - PubMed
-
- Rabinovich G. A., Ilarregui J. M. (2009) Immunol. Rev. 230, 144–159 - PubMed
-
- Rabinovich G. A., Toscano M. A. (2009) Nat. Rev. Immunol. 9, 338–352 - PubMed
-
- Salatino M., Croci D. O., Bianco G. A., Ilarregui J. M., Toscano M. A., Rabinovich G. A. (2008) Expert Opin. Biol. Ther. 8, 45–57 - PubMed
-
- Ley K., Kansas G. S. (2004) Nat. Rev. Immunol. 4, 325–335 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

