ADAM12 transmembrane and secreted isoforms promote breast tumor growth: a distinct role for ADAM12-S protein in tumor metastasis
- PMID: 21493715
- PMCID: PMC3121517
- DOI: 10.1074/jbc.M110.216036
ADAM12 transmembrane and secreted isoforms promote breast tumor growth: a distinct role for ADAM12-S protein in tumor metastasis
Abstract
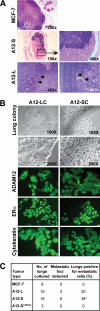
Increased levels of ADAM12 have been reported in a variety of human cancers. We have previously reported that urinary ADAM12 is predictive of disease status in breast cancer patients and that ADAM12 protein levels in urine increase with progression of disease. On the basis of these findings, the goal of this study was to elucidate the contribution of ADAM12 in breast tumor growth and progression. Overexpression of both the ADAM12-L (transmembrane) and ADAM12-S (secreted) isoforms in human breast tumor cells resulted in a significantly higher rate of tumor take and increased tumor size. Cells expressing the enzymatically inactive form of the secreted isoform, ADAM12-S, had tumor take rates and tumor volumes similar to those of wild-type cells, suggesting that the tumor-promoting activity of ADAM12-S was a function of its proteolytic activity. Of the two isoforms, only the secreted isoform, ADAM12-S, enhanced the ability of tumor cells to migrate and invade in vitro and resulted in a higher incidence of local and distant metastasis in vivo. This stimulatory effect of ADAM12-S on migration and invasion was dependent on its catalytic activity. Expression of both ADAM12 isoforms was found to be significantly elevated in human malignant breast tissue. Taken together, our results suggest that ADAM12 overexpression results in increased tumor take, tumor size, and metastasis in vivo. These findings suggest that ADAM12 may represent a potential therapeutic target in breast cancer.
Figures
Similar articles
-
ADAM12 induces estrogen-independence in breast cancer cells.Breast Cancer Res Treat. 2012 Feb;131(3):731-41. doi: 10.1007/s10549-011-1431-4. Epub 2011 Mar 9. Breast Cancer Res Treat. 2012. PMID: 21387162 Free PMC article.
-
ADAM12 produced by tumor cells rather than stromal cells accelerates breast tumor progression.Mol Cancer Res. 2011 Nov;9(11):1449-61. doi: 10.1158/1541-7786.MCR-11-0100. Epub 2011 Aug 29. Mol Cancer Res. 2011. PMID: 21875931 Free PMC article.
-
Phenotypic diversity of breast cancer-related mutations in metalloproteinase-disintegrin ADAM12.PLoS One. 2014 Mar 20;9(3):e92536. doi: 10.1371/journal.pone.0092536. eCollection 2014. PLoS One. 2014. PMID: 24651654 Free PMC article.
-
Targeting ADAM12 in human disease: head, body or tail?Curr Pharm Des. 2009;15(20):2300-10. doi: 10.2174/138161209788682389. Curr Pharm Des. 2009. PMID: 19601832 Review.
-
Cellular roles of ADAM12 in health and disease.Int J Biochem Cell Biol. 2008;40(9):1685-702. doi: 10.1016/j.biocel.2008.01.025. Epub 2008 Feb 1. Int J Biochem Cell Biol. 2008. PMID: 18342566 Review.
Cited by
-
ADAM-12 as a diagnostic marker for the proliferation, migration and invasion in patients with small cell lung cancer.PLoS One. 2014 Jan 21;9(1):e85936. doi: 10.1371/journal.pone.0085936. eCollection 2014. PLoS One. 2014. PMID: 24465799 Free PMC article.
-
A disintegrin and metalloproteinase-12 (ADAM12): function, roles in disease progression, and clinical implications.Biochim Biophys Acta. 2013 Oct;1830(10):4445-55. doi: 10.1016/j.bbagen.2013.05.011. Epub 2013 May 13. Biochim Biophys Acta. 2013. PMID: 23680494 Free PMC article. Review.
-
Notch increases the shedding of HB-EGF by ADAM12 to potentiate invadopodia formation in hypoxia.J Cell Biol. 2013 Apr 15;201(2):279-92. doi: 10.1083/jcb.201209151. J Cell Biol. 2013. PMID: 23589494 Free PMC article.
-
ADAM8 expression in invasive breast cancer promotes tumor dissemination and metastasis.EMBO Mol Med. 2014 Feb;6(2):278-94. doi: 10.1002/emmm.201303373. Epub 2013 Dec 27. EMBO Mol Med. 2014. PMID: 24375628 Free PMC article.
-
Adamalysines as Biomarkers and a Potential Target of Therapy in Colorectal Cancer Patients: Preliminary Results.Dis Markers. 2019 Sep 2;2019:5035234. doi: 10.1155/2019/5035234. eCollection 2019. Dis Markers. 2019. PMID: 31565100 Free PMC article.
References
-
- Gilpin B. J., Loechel F., Mattei M. G., Engvall E., Albrechtsen R., Wewer U. M. (1998) J. Biol. Chem. 273, 157–166 - PubMed
-
- Asakura M., Kitakaze M., Takashima S., Liao Y., Ishikura F., Yoshinaka T., Ohmoto H., Node K., Yoshino K., Ishiguro H., Asanuma H., Sanada S., Matsumura Y., Takeda H., Beppu S., Tada M., Hori M., Higashiyama S. (2002) Nat. Med. 8, 35–40 - PubMed
-
- Horiuchi K., Zhou H. M., Kelly K., Manova K., Blobel C. P. (2005) Dev. Biol. 283, 459–471 - PubMed
-
- Ito N., Nomura S., Iwase A., Ito T., Kikkawa F., Tsujimoto M., Ishiura S., Mizutani S. (2004) Biochem. Biophys. Res. Commun. 314, 1008–1013 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

