Rat mitochondrion-neuron focused microarray (rMNChip) and bioinformatics tools for rapid identification of differential pathways in brain tissues
- PMID: 21494430
- PMCID: PMC3076503
- DOI: 10.7150/ijbs.7.308
Rat mitochondrion-neuron focused microarray (rMNChip) and bioinformatics tools for rapid identification of differential pathways in brain tissues
Abstract
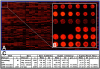
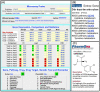
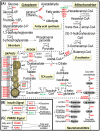
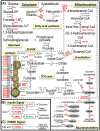
Mitochondrial function is of particular importance in brain because of its high demand for energy (ATP) and efficient removal of reactive oxygen species (ROS). We developed rat mitochondrion-neuron focused microarray (rMNChip) and integrated bioinformatics tools for rapid identification of differential pathways in brain tissues. rMNChip contains 1,500 genes involved in mitochondrial functions, stress response, circadian rhythms and signal transduction. The bioinformatics tool includes an algorithm for computing of differentially expressed genes, and a database for straightforward and intuitive interpretation for microarray results. Our application of these tools to RNA samples derived from rat frontal cortex (FC), hippocampus (HC) and hypothalamus (HT) led to the identification of differentially-expressed signal-transduction-bioenergenesis and neurotransmitter-synthesis pathways with a dominant number of genes (FC/HC = 55/6; FC/HT = 55/4) having significantly (p<0.05, FDR<10.70%) higher (≥1.25 fold) RNA levels in the frontal cortex than the others, strongly suggesting active generation of ATP and neurotransmitters and efficient removal of ROS. Thus, these tools for rapid and efficient identification of differential pathways in brain regions will greatly facilitate our systems-biological study and understanding of molecular mechanisms underlying complex and multifactorial neurodegenerative diseases.
Keywords: Microarray; bioinformatics; canonical pathway; mitochondrion; neuroscience.
Conflict of interest statement
Competing Interests: Authors state that there is no competing conflict of interests related to this publication.
Figures

Similar articles
-
Integrated bioinformatics analysis for differentially expressed genes and signaling pathways identification in gastric cancer.Int J Med Sci. 2021 Jan 1;18(3):792-800. doi: 10.7150/ijms.47339. eCollection 2021. Int J Med Sci. 2021. PMID: 33437215 Free PMC article.
-
BART: bioinformatics array research tool.BMC Bioinformatics. 2018 Aug 8;19(1):296. doi: 10.1186/s12859-018-2308-x. BMC Bioinformatics. 2018. PMID: 30089462 Free PMC article.
-
Bioinformatics Resource Manager v2.3: an integrated software environment for systems biology with microRNA and cross-species analysis tools.BMC Bioinformatics. 2012 Nov 23;13:311. doi: 10.1186/1471-2105-13-311. BMC Bioinformatics. 2012. PMID: 23174015 Free PMC article.
-
Bioinformatics analysis of microarray data.Methods Mol Biol. 2009;573:259-84. doi: 10.1007/978-1-60761-247-6_15. Methods Mol Biol. 2009. PMID: 19763933 Review.
-
DNA microarray technology and bioinformatic web services.Acta Microbiol Immunol Hung. 2019 Mar 1;66(1):19-30. doi: 10.1556/030.65.2018.028. Epub 2018 Jul 16. Acta Microbiol Immunol Hung. 2019. PMID: 30010394 Review.
Cited by
-
Biomarkers in an animal model for revealing neural, hematologic, and behavioral correlates of PTSD.J Vis Exp. 2012 Oct 10;(68):3361. doi: 10.3791/3361. J Vis Exp. 2012. PMID: 23093202 Free PMC article.
-
Mitochondrial Gene Expression Profiles and Metabolic Pathways in the Amygdala Associated with Exaggerated Fear in an Animal Model of PTSD.Front Neurol. 2014 Sep 23;5:164. doi: 10.3389/fneur.2014.00164. eCollection 2014. Front Neurol. 2014. PMID: 25295026 Free PMC article.
-
Mapping Molecular Datasets Back to the Brain Regions They are Extracted from: Remembering the Native Countries of Hypothalamic Expatriates and Refugees.Adv Neurobiol. 2018;21:101-193. doi: 10.1007/978-3-319-94593-4_6. Adv Neurobiol. 2018. PMID: 30334222 Free PMC article.
-
Sex-related differences and associated transcriptional signatures in the brain ventricular system and cerebrospinal fluid development in full-term neonates.Biol Sex Differ. 2025 May 25;16(1):35. doi: 10.1186/s13293-025-00719-2. Biol Sex Differ. 2025. PMID: 40414938 Free PMC article.
-
Automatic Spot Identification for High Throughput Microarray Analysis.J Bioeng Biomed Sci. 2011 Nov 18;Suppl 5:005. doi: 10.4172/2155-9538.S5-005. J Bioeng Biomed Sci. 2011. PMID: 24298393 Free PMC article.
References
-
- Zhang L, Zhou R, Li X, Ursano RJ, Li H. Stress-induced change of mitochondria membrane potential regulated by genomic and non-genomic GR signaling: a possible mechanism for hippocampus atrophy in PTSD. Med Hypotheses. 2006;66(6):1205–8. - PubMed
-
- Manoli I, Alesci S, Blackman MR, Su YA, Rennert OM, Chrousos GP. Mitochondria as key components of the stress response. Trends Endocrinol Metab. 2007 Jul;18(5):190–8. - PubMed
-
- Scheffler IE. A century of mitochondrial research: achievements and perspectives. Mitochondrion. 2001 Jun;1(1):3–31. - PubMed
-
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006 Jul 25;16(14):R551–R560. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

