Geographical and ethnic distribution of the HBV C/D recombinant on the Qinghai-Tibet Plateau
- PMID: 21494570
- PMCID: PMC3073994
- DOI: 10.1371/journal.pone.0018708
Geographical and ethnic distribution of the HBV C/D recombinant on the Qinghai-Tibet Plateau
Abstract
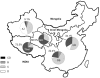
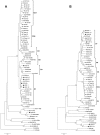
Two forms of hepatitis B virus (HBV) C/D recombinant have been identified in western China, but little is known about their geographical and ethnic distributions, and particularly the clinical significance and specific mutations in the pre-core region. To address these questions, a total of 624 chronic HBV carriers from four ethnic populations representing five provinces in western China were enrolled in this study. Genotypes were firstly determined by restriction fragment length polymorphism, and then confirmed by full or partial genome nucleotide sequencing. The distribution of HBV genotypes was as follows: HBV/B: 40 (6.4%); HBV/C: 221 (35.4%); HBV/D: 39 (6.3%); HBV/CD: 324 (51.9%). In the 324 HBV C/D recombinant infections, 244 (75.3%) were infected with the "CD1" and 80 (24.7%) were infected with the "CD2." The distribution of HBV genotypes exhibited distinct patterns in different regions and ethnic populations. Geographically, the C/D recombinant was the most prevalent HBV strain on the Qinghai-Tibet Plateau. Ethnically, the C/D recombinant had a higher prevalence in Tibetan patients than in other populations. Clinically, patients with HBV/CD1 showed significantly lower levels of serum total bilirubin than patients with HBV/C2. The prevalence of HBeAg was comparable between patients with HBV/CD1 and HBV/C2 (63.3% vs 50.0%, P = 0.118) whether patients were taken together or stratified by age into three groups (65.6% vs 58.8% in <30 years, P = 0.758; 61.9% vs 48.0% in 30-50 years, P = 0.244; 64.3% vs 33.3%, P = 0.336). Virologically HBV/CD1 had a significantly lower frequency of G1896A than HBV/C2. In conclusion, the HBV C/D recombinant is restricted to the Qinghai-Tibet Plateau in western China and is found predominantly in Tibetans. The predominance of the premature pre-core stop mutation G1896A in patients with the HBV C/D recombinant may account for the higher prevalence of HBeAg in these patients.
Conflict of interest statement
Figures
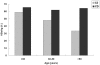

References
-
- Mulyanto, Depamede SN, Surayah K, Tjahyono AA, Jirintai, et al. Identification and characterization of novel hepatitis B virus subgenotype C10 in Nusa Tenggara, Indonesia. Arch Virol. 2010;155:705–715. - PubMed
-
- Luo K, Liu Z, He H, Peng J, Liang W, et al. The putative recombination of hepatitis B virus genotype B with pre-C/C region of genotype C. Virus Genes. 2004;29:31–41. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

