Genetic amplification of the vascular endothelial growth factor (VEGF) pathway genes, including VEGFA, in human osteosarcoma
- PMID: 21495021
- PMCID: PMC3465081
- DOI: 10.1002/cncr.26116
Genetic amplification of the vascular endothelial growth factor (VEGF) pathway genes, including VEGFA, in human osteosarcoma
Abstract
Background: Osteosarcoma is the most common primary tumor of bone. It is a highly vascular and extremely destructive malignancy that mainly affects children and young adults. The authors conducted microarray-based comparative genomic hybridization (aCGH) and pathway analyses to gain a systemic view of pathway alterations in the genetically altered genes.
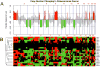
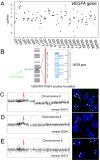
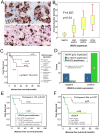
Methods: Recurrent amplified and deleted genes that were detected by aCGH were subjected to an analysis based on the Kyoto Encyclopedia of Genes and Genomes to identify the altered pathways. Among the enriched pathways, vascular endothelial growth factor (VEGF) pathway genes collectively were amplified, and alterations of this pathway were validated by fluorescence in situ hybridization (FISH) and immunohistochemistry analyses in 58 formalin-fixed, paraffin-embedded osteosarcoma archival tissues that had clinical follow-up information.
Results: The pathway enrichment analyses of the aCGH data revealed that VEGF pathway genes, including the VEGFA gene itself, were amplified significantly in osteosarcoma. Genetic amplification of the VEGFA gene, both focally and in larger fragment, was validated by FISH analysis. It is noteworthy that amplification of the VEGFA gene and elevated expression of the VEGFA protein were associated significantly with microvascular density and adverse tumor-free survival in patients with osteosarcoma.
Conclusions: The authors report for the first time that VEGF pathway genes, including the VEGFA gene, are amplified in osteosarcoma. Amplification of the VEGFA gene is not only an important mechanism for elevated VEGFA protein expression but also is a poor prognostic factor for tumor-free survival. Combined classification of VEGFA gene amplification and positive VEGFA protein expression may provide a more accurate stratification method of selecting anti-VEGF therapy for patients with osteosarcoma.
Copyright © 2011 American Cancer Society.
Figures

Similar articles
-
Correlation of WWOX, RUNX2 and VEGFA protein expression in human osteosarcoma.BMC Med Genomics. 2013 Dec 15;6:56. doi: 10.1186/1755-8794-6-56. BMC Med Genomics. 2013. PMID: 24330824 Free PMC article.
-
The genetic basis for inactivation of Wnt pathway in human osteosarcoma.BMC Cancer. 2014 Jun 18;14:450. doi: 10.1186/1471-2407-14-450. BMC Cancer. 2014. PMID: 24942472 Free PMC article.
-
VEGFA locus amplification potentially predicts a favorable prognosis in gastric adenocarcinoma.Pathol Res Pract. 2024 Aug;260:155441. doi: 10.1016/j.prp.2024.155441. Epub 2024 Jul 1. Pathol Res Pract. 2024. PMID: 38986362
-
Correlation of vascular endothelial growth factor with survival and pathological characteristics of patients with osteosarcoma: A systematic review and meta-analysis.Eur J Cancer Care (Engl). 2022 Sep;31(5):e13629. doi: 10.1111/ecc.13629. Epub 2022 Jun 16. Eur J Cancer Care (Engl). 2022. PMID: 35707976
-
Intracranial meningiomas, the VEGF-A pathway, and peritumoral brain oedema.Dan Med J. 2013 Apr;60(4):B4626. Dan Med J. 2013. PMID: 23651727 Review.
Cited by
-
Cyr61 silencing reduces vascularization and dissemination of osteosarcoma tumors.Oncogene. 2015 Jun 11;34(24):3207-13. doi: 10.1038/onc.2014.232. Epub 2014 Jul 28. Oncogene. 2015. PMID: 25065593
-
The Protein Tyrosine Phosphatase Rptpζ Suppresses Osteosarcoma Development in Trp53-Heterozygous Mice.PLoS One. 2015 Sep 11;10(9):e0137745. doi: 10.1371/journal.pone.0137745. eCollection 2015. PLoS One. 2015. PMID: 26360410 Free PMC article.
-
Real-World Data on Cabozantinib in Advanced Osteosarcoma and Ewing Sarcoma Patients: A Study from the Hellenic Group of Sarcoma and Rare Cancers.J Clin Med. 2023 Jan 31;12(3):1119. doi: 10.3390/jcm12031119. J Clin Med. 2023. PMID: 36769769 Free PMC article.
-
Correlation of WWOX, RUNX2 and VEGFA protein expression in human osteosarcoma.BMC Med Genomics. 2013 Dec 15;6:56. doi: 10.1186/1755-8794-6-56. BMC Med Genomics. 2013. PMID: 24330824 Free PMC article.
-
Transforming Growth Factor-β Signaling Plays a Pivotal Role in the Interplay Between Osteosarcoma Cells and Their Microenvironment.Front Oncol. 2018 Apr 30;8:133. doi: 10.3389/fonc.2018.00133. eCollection 2018. Front Oncol. 2018. PMID: 29761075 Free PMC article. Review.
References
-
- Unni K Krishnan, I CY. Dahlin's bone tumours. Sixth. Vol. 123 Philadelphia: Lippincott Williams & Wilkins; 2010.
-
- Yang J, Cogdell D, Yang D, Hu L, Li H, Zheng H, et al. Deletion of the WWOX gene and frequent loss of its protein expression in human osteosarcoma. Cancer Lett. 291(1):31–8. - PubMed
-
- Quan GM, Choong PF. Anti-angiogenic therapy for osteosarcoma. Cancer Metastasis Rev. 2006;25(4):707–13. - PubMed
-
- Dvorak HF. Vascular permeability factor/vascular endothelial growth factor: a critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. J Clin Oncol. 2002;20(21):4368–80. - PubMed
-
- Zangari M, Anaissie E, Stopeck A, Morimoto A, Tan N, Lancet J, et al. Phase II study of SU5416, a small molecule vascular endothelial growth factor tyrosine kinase receptor inhibitor, in patients with refractory multiple myeloma. Clin Cancer Res. 2004;10(1 Pt 1):88–95. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

