SNP-based pathway enrichment analysis for genome-wide association studies
- PMID: 21496265
- PMCID: PMC3102637
- DOI: 10.1186/1471-2105-12-99
SNP-based pathway enrichment analysis for genome-wide association studies
Abstract
Background: Recently we have witnessed a surge of interest in using genome-wide association studies (GWAS) to discover the genetic basis of complex diseases. Many genetic variations, mostly in the form of single nucleotide polymorphisms (SNPs), have been identified in a wide spectrum of diseases, including diabetes, cancer, and psychiatric diseases. A common theme arising from these studies is that the genetic variations discovered by GWAS can only explain a small fraction of the genetic risks associated with the complex diseases. New strategies and statistical approaches are needed to address this lack of explanation. One such approach is the pathway analysis, which considers the genetic variations underlying a biological pathway, rather than separately as in the traditional GWAS studies. A critical challenge in the pathway analysis is how to combine evidences of association over multiple SNPs within a gene and multiple genes within a pathway. Most current methods choose the most significant SNP from each gene as a representative, ignoring the joint action of multiple SNPs within a gene. This approach leads to preferential identification of genes with a greater number of SNPs.
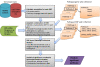
Results: We describe a SNP-based pathway enrichment method for GWAS studies. The method consists of the following two main steps: 1) for a given pathway, using an adaptive truncated product statistic to identify all representative (potentially more than one) SNPs of each gene, calculating the average number of representative SNPs for the genes, then re-selecting the representative SNPs of genes in the pathway based on this number; and 2) ranking all selected SNPs by the significance of their statistical association with a trait of interest, and testing if the set of SNPs from a particular pathway is significantly enriched with high ranks using a weighted Kolmogorov-Smirnov test. We applied our method to two large genetically distinct GWAS data sets of schizophrenia, one from European-American (EA) and the other from African-American (AA). In the EA data set, we found 22 pathways with nominal P-value less than or equal to 0.001 and corresponding false discovery rate (FDR) less than 5%. In the AA data set, we found 11 pathways by controlling the same nominal P-value and FDR threshold. Interestingly, 8 of these pathways overlap with those found in the EA sample. We have implemented our method in a JAVA software package, called SNP Set Enrichment Analysis (SSEA), which contains a user-friendly interface and is freely available at http://cbcl.ics.uci.edu/SSEA.
Conclusions: The SNP-based pathway enrichment method described here offers a new alternative approach for analysing GWAS data. By applying it to schizophrenia GWAS studies, we show that our method is able to identify statistically significant pathways, and importantly, pathways that can be replicated in large genetically distinct samples.
Figures
Similar articles
-
Genome-wide genetic analyses highlight mitogen-activated protein kinase (MAPK) signaling in the pathogenesis of endometriosis.Hum Reprod. 2017 Apr 1;32(4):780-793. doi: 10.1093/humrep/dex024. Hum Reprod. 2017. PMID: 28333195 Free PMC article.
-
Pathway analysis of a genome-wide association study in schizophrenia.Gene. 2013 Aug 1;525(1):107-15. doi: 10.1016/j.gene.2013.04.014. Epub 2013 May 1. Gene. 2013. PMID: 23644028
-
Pathway analysis of genome-wide association studies for Parkinson's disease.Mol Biol Rep. 2013 Mar;40(3):2599-607. doi: 10.1007/s11033-012-2346-9. Epub 2012 Dec 13. Mol Biol Rep. 2013. PMID: 23238920
-
Shared genetic etiology underlying Alzheimer's disease and type 2 diabetes.Mol Aspects Med. 2015 Jun-Oct;43-44:66-76. doi: 10.1016/j.mam.2015.06.006. Epub 2015 Jun 23. Mol Aspects Med. 2015. PMID: 26116273 Free PMC article. Review.
-
Genomewide association studies: history, rationale, and prospects for psychiatric disorders.Am J Psychiatry. 2009 May;166(5):540-56. doi: 10.1176/appi.ajp.2008.08091354. Epub 2009 Apr 1. Am J Psychiatry. 2009. PMID: 19339359 Free PMC article. Review.
Cited by
-
Between candidate genes and whole genomes: time for alternative approaches in blood pressure genetics.Curr Hypertens Rep. 2012 Feb;14(1):46-61. doi: 10.1007/s11906-011-0241-8. Curr Hypertens Rep. 2012. PMID: 22161147 Free PMC article. Review.
-
Maximal information component analysis: a novel non-linear network analysis method.Front Genet. 2013 Mar 12;4:28. doi: 10.3389/fgene.2013.00028. eCollection 2013. Front Genet. 2013. PMID: 23487572 Free PMC article.
-
Predicting disease risk using bootstrap ranking and classification algorithms.PLoS Comput Biol. 2013;9(8):e1003200. doi: 10.1371/journal.pcbi.1003200. Epub 2013 Aug 22. PLoS Comput Biol. 2013. PMID: 23990773 Free PMC article.
-
Several Critical Cell Types, Tissues, and Pathways Are Implicated in Genome-Wide Association Studies for Systemic Lupus Erythematosus.G3 (Bethesda). 2016 Jun 1;6(6):1503-11. doi: 10.1534/g3.116.027326. G3 (Bethesda). 2016. PMID: 27172182 Free PMC article.
-
Candidate gene-environment interaction research: reflections and recommendations.Perspect Psychol Sci. 2015 Jan;10(1):37-59. doi: 10.1177/1745691614556682. Perspect Psychol Sci. 2015. PMID: 25620996 Free PMC article. Review.
References
-
- Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JRB, Rayner NW, Freathy RM. et al.Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science (New York, NY) 2007;316:1336–1341. doi: 10.1126/science.1142364. - DOI - PMC - PubMed
-
- Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson Ba, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M. et al.Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nature genetics. 2009;41:1122–1126. doi: 10.1038/ng.448. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases