The sequence, structure and evolutionary features of HOTAIR in mammals
- PMID: 21496275
- PMCID: PMC3103462
- DOI: 10.1186/1471-2148-11-102
The sequence, structure and evolutionary features of HOTAIR in mammals
Abstract
Background: An increasing number of long noncoding RNAs (lncRNAs) have been identified recently. Different from all the others that function in cis to regulate local gene expression, the newly identified HOTAIR is located between HoxC11 and HoxC12 in the human genome and regulates HoxD expression in multiple tissues. Like the well-characterised lncRNA Xist, HOTAIR binds to polycomb proteins to methylate histones at multiple HoxD loci, but unlike Xist, many details of its structure and function, as well as the trans regulation, remain unclear. Moreover, HOTAIR is involved in the aberrant regulation of gene expression in cancer.
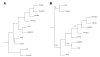
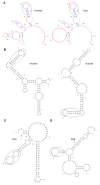
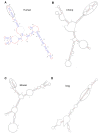
Results: To identify conserved domains in HOTAIR and study the phylogenetic distribution of this lncRNA, we searched the genomes of 10 mammalian and 3 non-mammalian vertebrates for matches to its 6 exons and the two conserved domains within the 1800 bp exon6 using Infernal. There was just one high-scoring hit for each mammal, but many low-scoring hits were found in both mammals and non-mammalian vertebrates. These hits and their flanking genes in four placental mammals and platypus were examined to determine whether HOTAIR contained elements shared by other lncRNAs. Several of the hits were within unknown transcripts or ncRNAs, many were within introns of, or antisense to, protein-coding genes, and conservation of the flanking genes was observed only between human and chimpanzee. Phylogenetic analysis revealed discrete evolutionary dynamics for orthologous sequences of HOTAIR exons. Exon1 at the 5' end and a domain in exon6 near the 3' end, which contain domains that bind to multiple proteins, have evolved faster in primates than in other mammals. Structures were predicted for exon1, two domains of exon6 and the full HOTAIR sequence. The sequence and structure of two fragments, in exon1 and the domain B of exon6 respectively, were identified to robustly occur in predicted structures of exon1, domain B of exon6 and the full HOTAIR in mammals.
Conclusions: HOTAIR exists in mammals, has poorly conserved sequences and considerably conserved structures, and has evolved faster than nearby HoxC genes. Exons of HOTAIR show distinct evolutionary features, and a 239 bp domain in the 1804 bp exon6 is especially conserved. These features, together with the absence of some exons and sequences in mouse, rat and kangaroo, suggest ab initio generation of HOTAIR in marsupials. Structure prediction identifies two fragments in the 5' end exon1 and the 3' end domain B of exon6, with sequence and structure invariably occurring in various predicted structures of exon1, the domain B of exon6 and the full HOTAIR.
Figures
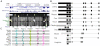
Similar articles
-
Structural and functional differences in the long non-coding RNA hotair in mouse and human.PLoS Genet. 2011 May;7(5):e1002071. doi: 10.1371/journal.pgen.1002071. Epub 2011 May 26. PLoS Genet. 2011. PMID: 21637793 Free PMC article.
-
Long noncoding RNA in genome regulation: prospects and mechanisms.RNA Biol. 2010 Sep-Oct;7(5):582-5. doi: 10.4161/rna.7.5.13216. Epub 2010 Sep 1. RNA Biol. 2010. PMID: 20930520 Free PMC article.
-
The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene.Science. 2006 Jun 16;312(5780):1653-5. doi: 10.1126/science.1126316. Science. 2006. PMID: 16778056
-
The four dimensions of noncoding RNA conservation.Trends Genet. 2014 Apr;30(4):121-3. doi: 10.1016/j.tig.2014.01.004. Epub 2014 Mar 7. Trends Genet. 2014. PMID: 24613441 Review.
-
Evolution and Coevolution of PRC2 Genes in Vertebrates and Mammals.Adv Protein Chem Struct Biol. 2015;101:125-48. doi: 10.1016/bs.apcsb.2015.06.010. Epub 2015 Aug 4. Adv Protein Chem Struct Biol. 2015. PMID: 26572978 Review.
Cited by
-
Functions and underlying mechanisms of lncRNA HOTAIR in cancer chemotherapy resistance.Cell Death Discov. 2022 Sep 13;8(1):383. doi: 10.1038/s41420-022-01174-3. Cell Death Discov. 2022. PMID: 36100611 Free PMC article. Review.
-
Roles of lncRNA in breast cancer.Front Biosci (Schol Ed). 2015 Jun 1;7(1):94-108. doi: 10.2741/S427. Front Biosci (Schol Ed). 2015. PMID: 25961689 Free PMC article. Review.
-
Large intervening non-coding RNA HOTAIR is an indicator of poor prognosis and a therapeutic target in human cancers.Int J Mol Sci. 2014 Oct 20;15(10):18985-99. doi: 10.3390/ijms151018985. Int J Mol Sci. 2014. PMID: 25334066 Free PMC article. Review.
-
The functional role of long non-coding RNAs and epigenetics.Biol Proced Online. 2014 Sep 15;16:11. doi: 10.1186/1480-9222-16-11. eCollection 2014. Biol Proced Online. 2014. PMID: 25276098 Free PMC article. Review.
-
The Role of HOTAIR/miR-148b-3p/USF1 on Regulating the Permeability of BTB.Front Mol Neurosci. 2017 Jun 28;10:194. doi: 10.3389/fnmol.2017.00194. eCollection 2017. Front Mol Neurosci. 2017. Retraction in: Front Mol Neurosci. 2023 May 02;16:1207936. doi: 10.3389/fnmol.2023.1207936. PMID: 28701916 Free PMC article. Retracted.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

