Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine
- PMID: 21496645
- PMCID: PMC3108460
- DOI: 10.1016/j.cell.2011.03.036
Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine
Abstract
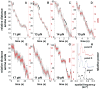
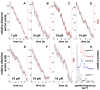
All cells employ ATP-powered proteases for protein-quality control and regulation. In the ClpXP protease, ClpX is a AAA+ machine that recognizes specific protein substrates, unfolds these molecules, and then translocates the denatured polypeptide through a central pore and into ClpP for degradation. Here, we use optical-trapping nanometry to probe the mechanics of enzymatic unfolding and translocation of single molecules of a multidomain substrate. Our experiments demonstrate the capacity of ClpXP and ClpX to perform mechanical work under load, reveal very fast and highly cooperative unfolding of individual substrate domains, suggest a translocation step size of 5-8 amino acids, and support a power-stroke model of denaturation in which successful enzyme-mediated unfolding of stable domains requires coincidence between mechanical pulling by the enzyme and a transient stochastic reduction in protein stability. We anticipate that single-molecule studies of the mechanical properties of other AAA+ proteolytic machines will reveal many shared features with ClpXP.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures




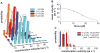
Comment in
-
Protease power strokes force proteins to unfold.Cell. 2011 Apr 29;145(3):339-40. doi: 10.1016/j.cell.2011.04.007. Cell. 2011. PMID: 21529709 Free PMC article.
References
-
- Burton BM, Williams TL, Baker TA. ClpX-mediated remodeling of mu transpososomes: selective unfolding of subunits destabilizes the entire complex. Mol Cell. 2001;8:449–454. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

