Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1
- PMID: 21497082
- PMCID: PMC3146975
- DOI: 10.1016/j.gde.2011.03.002
Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1
Abstract
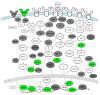
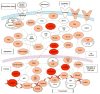
The GU-rich element (GRE) was identified as a conserved sequence enriched in the 3' UTR of human transcripts that exhibited rapid mRNA turnover. In mammalian cells, binding to GREs by the protein CELF1 coordinates mRNA decay of networks of transcripts involved in cell growth, migration, and apoptosis. Depending on the context, GREs and CELF1 also regulate pre-mRNA splicing and translation. GREs are highly conserved throughout evolution and play important roles in the development of organisms ranging from worms to man. In humans, abnormal GRE-mediated regulation contributes to disease states and cancer. Thus, GREs and CELF proteins serve critical functions in gene expression regulation and define an important evolutionarily conserved posttranscriptional regulatory network.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- Mansfield KD, Keene JD. The ribonome: a dominant force in co-ordinating gene expression. Biol Cell. 2009;101(3):169–181. This review discusses the post-transcriptional RNA operon theory of co-regulated gene expression, whereby the coordinated dynamics of RNA-binding proteins allows for the recombination and remodelling of the RNPs (ribonucleoproteins) to generate new combinations of functionally related proteins and keep the global mRNA environment in balance. - PMC - PubMed
-
- Keene JD. Biological clocks and the coordination theory of RNA operons and regulons. Cold Spring Harb Symp Quant Biol. 2007;72:157–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

