Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division
- PMID: 21497118
- PMCID: PMC3088519
- DOI: 10.1016/j.immuni.2011.03.017
Asymmetric proteasome segregation as a mechanism for unequal partitioning of the transcription factor T-bet during T lymphocyte division
Abstract
Polarized segregation of proteins in T cells is thought to play a role in diverse cellular functions including signal transduction, migration, and directed secretion of cytokines. Persistence of this polarization can result in asymmetric segregation of fate-determining proteins during cell division, which may enable a T cell to generate diverse progeny. Here, we provide evidence that a lineage-determining transcription factor, T-bet, underwent asymmetric organization in activated T cells preparing to divide and that it was unequally partitioned into the two daughter cells. This unequal acquisition of T-bet appeared to result from its asymmetric destruction during mitosis by virtue of concomitant asymmetric segregation of the proteasome. These results suggest a mechanism by which a cell may unequally localize cellular activities during division, thereby imparting disparity in the abundance of cell fate regulators in the daughter cells.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures
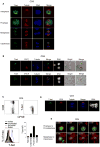
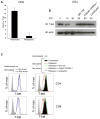

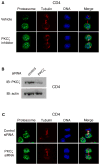


Comment in
-
T cells: T cell fate in the (im)balance.Nat Rev Immunol. 2011 Jun;11(6):367. doi: 10.1038/nri2995. Epub 2011 May 20. Nat Rev Immunol. 2011. PMID: 21597476 No abstract available.
References
-
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat Immunol. 2002;3:549–557. - PubMed
-
- Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. - PubMed
-
- Berg LJ. Signalling through TEC kinases regulates conventional versus innate CD8(+) T-cell development. Nat Rev Immunol. 2007;7:479–485. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K01 AR052802/AR/NIAMS NIH HHS/United States
- R01 AI037584/AI/NIAID NIH HHS/United States
- R56 AI076458/AI/NIAID NIH HHS/United States
- R01 AI084987/AI/NIAID NIH HHS/United States
- R37 GM053256/GM/NIGMS NIH HHS/United States
- R01 AI042370/AI/NIAID NIH HHS/United States
- AR052802/AR/NIAMS NIH HHS/United States
- R01 AI046564/AI/NIAID NIH HHS/United States
- GM053256/GM/NIGMS NIH HHS/United States
- AI061699/AI/NIAID NIH HHS/United States
- R01 AI076458/AI/NIAID NIH HHS/United States
- AI37584/AI/NIAID NIH HHS/United States
- T32 AI055428/AI/NIAID NIH HHS/United States
- R24 DK080506/DK/NIDDK NIH HHS/United States
- DK80506/DK/NIDDK NIH HHS/United States
- P01 CA093615/CA/NCI NIH HHS/United States
- T32 HD007516/HD/NICHD NIH HHS/United States
- K08 DK080949/DK/NIDDK NIH HHS/United States
- R01 GM053256/GM/NIGMS NIH HHS/United States
- T32HD007516/HD/NICHD NIH HHS/United States
- DK080949/DK/NIDDK NIH HHS/United States
- AI46564/AI/NIAID NIH HHS/United States
- CA093615/CA/NCI NIH HHS/United States
- AI076458/AI/NIAID NIH HHS/United States
- R01 AI061699/AI/NIAID NIH HHS/United States
- AI042370/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

