Regulation of γ-globin gene expression involves signaling through the p38 MAPK/CREB1 pathway
- PMID: 21497119
- PMCID: PMC3695476
- DOI: 10.1016/j.bcmd.2011.03.003
Regulation of γ-globin gene expression involves signaling through the p38 MAPK/CREB1 pathway
Abstract
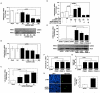
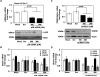
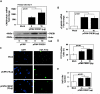
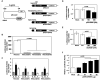
In response to sodium butyrate and trichostatin A treatment in erythroid cells, p38 mitogen activated protein kinase (MAPK) mediates fetal hemoglobin (HbF) induction by activating cAMP response element binding protein 1 (CREB1). To expand on this observation, we completed studies to determine the role of p38 MAPK in steady-state γ-globin regulation. We propose that p38 signaling regulates Gγ-globin transcription during erythroid maturation through its downstream effector CREB1 which binds the Gγ-globin cAMP response element (G-CRE). We demonstrated that a loss of p38 or CREB1 function by siRNA knockdown resulted in target gene silencing. Moreover, gain of p38 or CREB1 function augments γ-globin transcription. These regulatory effects were conserved under physiological conditions tested in primary erythroid cells. When the G-CRE was mutated in a stable chromatin environment Gγ-globin promoter activity was nearly abolished. Furthermore, introduction of mutations in the G-CRE abolished Gγ-globin activation via p38 MAPK/CREB1 signaling. Chromatin immunoprecipitation assays (ChIP) demonstrated that CREB1 and its binding partner CREB binding protein (CBP) co-localize at the G-CRE region. These data support the role of p38 MAPK/CREB1 signaling in Gγ-globin gene transcription under steady-state conditions.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Regulation of Gγ-globin gene by ATF2 and its associated proteins through the cAMP-response element.PLoS One. 2013 Nov 6;8(11):e78253. doi: 10.1371/journal.pone.0078253. eCollection 2013. PLoS One. 2013. PMID: 24223142 Free PMC article.
-
cJun modulates Ggamma-globin gene expression via an upstream cAMP response element.Blood Cells Mol Dis. 2010 Jan 15;44(1):7-15. doi: 10.1016/j.bcmd.2009.10.002. Epub 2009 Oct 27. Blood Cells Mol Dis. 2010. PMID: 19861239 Free PMC article.
-
Oridonin enhances γ‑globin expression in erythroid precursors from patients with β‑thalassemia via activation of p38 MAPK signaling.Mol Med Rep. 2020 Feb;21(2):909-917. doi: 10.3892/mmr.2019.10848. Epub 2019 Nov 25. Mol Med Rep. 2020. PMID: 31789406
-
Mechanism for fetal hemoglobin induction by histone deacetylase inhibitors involves gamma-globin activation by CREB1 and ATF-2.Blood. 2006 Nov 15;108(10):3590-9. doi: 10.1182/blood-2006-01-023713. Epub 2006 Aug 8. Blood. 2006. PMID: 16896160 Free PMC article.
-
Evaluation of Signaling Pathways Involved in γ-Globin Gene Induction Using Fetal Hemoglobin Inducer Drugs.Int J Hematol Oncol Stem Cell Res. 2013;7(3):41-6. Int J Hematol Oncol Stem Cell Res. 2013. PMID: 24505534 Free PMC article. Review.
Cited by
-
Synthesis and pharmacological evaluation of pomalidomide derivatives useful for sickle cell disease treatment.Bioorg Chem. 2021 Sep;114:105077. doi: 10.1016/j.bioorg.2021.105077. Epub 2021 Jun 10. Bioorg Chem. 2021. PMID: 34130111 Free PMC article.
-
Original Research: A case-control genome-wide association study identifies genetic modifiers of fetal hemoglobin in sickle cell disease.Exp Biol Med (Maywood). 2016 Apr;241(7):706-18. doi: 10.1177/1535370216642047. Epub 2016 Mar 27. Exp Biol Med (Maywood). 2016. PMID: 27022141 Free PMC article.
-
Regulation of Gγ-globin gene by ATF2 and its associated proteins through the cAMP-response element.PLoS One. 2013 Nov 6;8(11):e78253. doi: 10.1371/journal.pone.0078253. eCollection 2013. PLoS One. 2013. PMID: 24223142 Free PMC article.
-
Globin gene expression in correlation with G protein-related genes during erythroid differentiation.BMC Genomics. 2013 Feb 20;14:116. doi: 10.1186/1471-2164-14-116. BMC Genomics. 2013. PMID: 23425329 Free PMC article.
-
Transmembrane Protein ANTXR1 Regulates γ-Globin Expression by Targeting the Wnt/β-Catenin Signaling Pathway.J Immunol Res. 2022 Jul 30;2022:8440422. doi: 10.1155/2022/8440422. eCollection 2022. J Immunol Res. 2022. PMID: 35942209 Free PMC article.
References
-
- Neiman AM. Conservation and reiteration of a kinase cascade. Trends Genet. 1993;9:390–4. - PubMed
-
- Nishida E, Gotoh Y. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem Sci. 1993;18:128–31. - PubMed
-
- Martin-Blanco E. p38 MAPK signalling cascades: ancient roles and new functions. Bioessays. 2000;22:637–45. - PubMed
-
- Steinbrech DS, Mehrara BJ, Saadeh PB, et al. VEGF expression in an osteoblast-like cell line is regulated by a hypoxia response mechanism. Am J Physiol Cell Physiol. 2000;278:C853–60. - PubMed
-
- Tamura K, Sudo T, Senftleben U, et al. Requirement for p38alpha in erythropoietin expression: a role for stress kinases in erythropoiesis. Cell. 2000;102:221–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

