Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx
- PMID: 21497756
- PMCID: PMC3086024
- DOI: 10.1016/j.devcel.2011.03.012
Pancreatic β cell identity is maintained by DNA methylation-mediated repression of Arx
Abstract
Adult pancreatic β cells can replicate during growth and after injury to maintain glucose homeostasis. Here, we report that β cells deficient in Dnmt1, an enzyme that propagates DNA methylation patterns during cell division, were converted to α cells. We identified the lineage determination gene aristaless-related homeobox (Arx), as methylated and repressed in β cells, and hypomethylated and expressed in α cells and Dnmt1-deficient β cells. We show that the methylated region of the Arx locus in β cells was bound by methyl-binding protein MeCP2, which recruited PRMT6, an enzyme that methylates histone H3R2 resulting in repression of Arx. This suggests that propagation of DNA methylation during cell division also ensures recruitment of enzymatic machinery capable of modifying and transmitting histone marks. Our results reveal that propagation of DNA methylation during cell division is essential for repression of α cell lineage determination genes to maintain pancreatic β cell identity.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


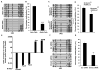
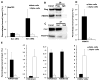

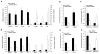
Comment in
-
Removing the brakes on cell identity.Dev Cell. 2011 Apr 19;20(4):411-2. doi: 10.1016/j.devcel.2011.04.002. Dev Cell. 2011. PMID: 21497752
References
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Collombat P, Hecksher-Sorensen J, Broccoli V, Krull J, Ponte I, Mundiger T, Smith J, Gruss P, Serup P, Mansouri A. The simultaneous loss of Arx and Pax4 genes promotes a somatostatin-producing cell fate specification at the expense of the alpha- and beta-cell lineages in the mouse endocrine pancreas. Development. 2005;132:2969–2980. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

