Escherichia coli thioredoxin-like protein YbbN contains an atypical tetratricopeptide repeat motif and is a negative regulator of GroEL
- PMID: 21498507
- PMCID: PMC3103325
- DOI: 10.1074/jbc.M111.238741
Escherichia coli thioredoxin-like protein YbbN contains an atypical tetratricopeptide repeat motif and is a negative regulator of GroEL
Abstract
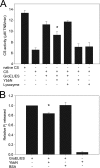
Many proteins contain a thioredoxin (Trx)-like domain fused with one or more partner domains that diversify protein function by the modular construction of new molecules. The Escherichia coli protein YbbN is a Trx-like protein that contains a C-terminal domain with low homology to tetratricopeptide repeat motifs. YbbN has been proposed to act as a chaperone or co-chaperone that aids in heat stress response and DNA synthesis. We report the crystal structure of YbbN, which is an elongated molecule with a mobile Trx domain and four atypical tetratricopeptide repeat motifs. The Trx domain lacks a canonical CXXC active site architecture and is not a functional oxidoreductase. A variety of proteins in E. coli interact with YbbN, including multiple ribosomal protein subunits and a strong interaction with GroEL. YbbN acts as a mild inhibitor of GroESL chaperonin function and ATPase activity, suggesting that it is a negative regulator of the GroESL system. Combined with previous observations that YbbN enhances the DnaK-DnaJ-GrpE chaperone system, we propose that YbbN coordinately regulates the activities of these two prokaryotic chaperones, thereby helping to direct client protein traffic initially to DnaK. Therefore, YbbN may play a role in integrating the activities of different chaperone pathways in E. coli and related bacteria.
Figures


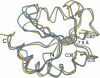



Similar articles
-
CnoX Is a Chaperedoxin: A Holdase that Protects Its Substrates from Irreversible Oxidation.Mol Cell. 2018 May 17;70(4):614-627.e7. doi: 10.1016/j.molcel.2018.04.002. Epub 2018 May 10. Mol Cell. 2018. PMID: 29754824
-
The thioredoxin homolog YbbN functions as a chaperone rather than as an oxidoreductase.Biochem Biophys Res Commun. 2008 Oct 3;374(4):668-72. doi: 10.1016/j.bbrc.2008.07.080. Epub 2008 Jul 25. Biochem Biophys Res Commun. 2008. PMID: 18657513
-
The Escherichia coli thioredoxin homolog YbbN/Trxsc is a chaperone and a weak protein oxidoreductase.Biochem Biophys Res Commun. 2006 May 12;343(3):780-6. doi: 10.1016/j.bbrc.2006.03.028. Epub 2006 Mar 20. Biochem Biophys Res Commun. 2006. PMID: 16563353
-
Interferon-gamma is a target for binding and folding by both Escherichia coli chaperone model systems GroEL/GroES and DnaK/DnaJ/GrpE.Biochimie. 1998 Aug-Sep;80(8-9):729-37. doi: 10.1016/s0300-9084(99)80026-1. Biochimie. 1998. PMID: 9865495 Review.
-
Recent advances in understanding catalysis of protein folding by molecular chaperones.FEBS Lett. 2020 Sep;594(17):2770-2781. doi: 10.1002/1873-3468.13844. Epub 2020 Jun 12. FEBS Lett. 2020. PMID: 32446288 Review.
Cited by
-
The Chaperone and Redox Properties of CnoX Chaperedoxins Are Tailored to the Proteostatic Needs of Bacterial Species.mBio. 2018 Nov 27;9(6):e01541-18. doi: 10.1128/mBio.01541-18. mBio. 2018. PMID: 30482828 Free PMC article.
-
Deciphering the Role of Multiple Thioredoxin Fold Proteins of Leptospirillum sp. in Oxidative Stress Tolerance.Int J Mol Sci. 2020 Mar 10;21(5):1880. doi: 10.3390/ijms21051880. Int J Mol Sci. 2020. PMID: 32164170 Free PMC article.
-
A molecular device for the redox quality control of GroEL/ES substrates.Cell. 2023 Mar 2;186(5):1039-1049.e17. doi: 10.1016/j.cell.2023.01.013. Epub 2023 Feb 9. Cell. 2023. PMID: 36764293 Free PMC article.
-
Fort CnoX: Protecting Bacterial Proteins From Misfolding and Oxidative Damage.Front Mol Biosci. 2021 May 4;8:681932. doi: 10.3389/fmolb.2021.681932. eCollection 2021. Front Mol Biosci. 2021. PMID: 34017858 Free PMC article. Review.
-
RcsB contributes to the distinct stress fitness among Escherichia coli O157:H7 curli variants of the 1993 hamburger-associated outbreak strains.Appl Environ Microbiol. 2012 Nov;78(21):7706-19. doi: 10.1128/AEM.02157-12. Epub 2012 Aug 24. Appl Environ Microbiol. 2012. PMID: 22923406 Free PMC article.
References
-
- Holmgren A. (1985) Annu. Rev. Biochem. 54, 237–271 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

