Identification of a novel muscle A-type lamin-interacting protein (MLIP)
- PMID: 21498514
- PMCID: PMC3103349
- DOI: 10.1074/jbc.M110.165548
Identification of a novel muscle A-type lamin-interacting protein (MLIP)
Abstract
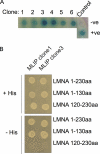
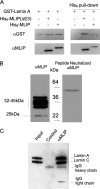
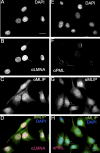
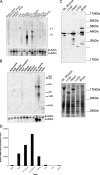
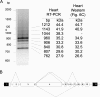
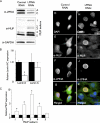
Mutations in the A-type lamin (LMNA) gene are associated with age-associated degenerative disorders of mesenchymal tissues, such as dilated cardiomyopathy, Emery-Dreifuss muscular dystrophy, and limb-girdle muscular dystrophy. The molecular mechanisms that connect mutations in LMNA with different human diseases are poorly understood. Here, we report the identification of a Muscle-enriched A-type Lamin-interacting Protein, MLIP (C6orf142 and 2310046A06rik), a unique single copy gene that is an innovation of amniotes (reptiles, birds, and mammals). MLIP encodes alternatively spliced variants (23-57 kDa) and possesses several novel structural motifs not found in other proteins. MLIP is expressed ubiquitously and most abundantly in heart, skeletal, and smooth muscle. MLIP interacts directly and co-localizes with lamin A and C in the nuclear envelope. MLIP also co-localizes with promyelocytic leukemia (PML) bodies within the nucleus. PML, like MLIP, is only found in amniotes, suggesting that a functional link between the nuclear envelope and PML bodies may exist through MLIP. Down-regulation of lamin A/C expression by shRNA results in the up-regulation and mislocalization of MLIP. Given that MLIP is expressed most highly in striated and smooth muscle, it is likely to contribute to the mesenchymal phenotypes of laminopathies.
Figures



Similar articles
-
Expression of murine muscle-enriched A-type lamin-interacting protein (MLIP) is regulated by tissue-specific alternative transcription start sites.J Biol Chem. 2018 Dec 21;293(51):19761-19770. doi: 10.1074/jbc.RA118.003758. Epub 2018 Nov 2. J Biol Chem. 2018. PMID: 30389785 Free PMC article.
-
Emery-Dreifuss Muscular Dystrophy-Associated Mutant Forms of Lamin A Recruit the Stress Responsive Protein Ankrd2 into the Nucleus, Affecting the Cellular Response to Oxidative Stress.Cell Physiol Biochem. 2017;42(1):169-184. doi: 10.1159/000477309. Epub 2017 May 25. Cell Physiol Biochem. 2017. PMID: 28531892
-
Emery-Dreifuss muscular dystrophy: focal point nuclear envelope.Curr Opin Neurol. 2019 Oct;32(5):728-734. doi: 10.1097/WCO.0000000000000741. Curr Opin Neurol. 2019. PMID: 31460960 Free PMC article. Review.
-
Lamin-Related Congenital Muscular Dystrophy Alters Mechanical Signaling and Skeletal Muscle Growth.Int J Mol Sci. 2020 Dec 30;22(1):306. doi: 10.3390/ijms22010306. Int J Mol Sci. 2020. PMID: 33396724 Free PMC article.
-
Laminopathies affecting skeletal and cardiac muscles: clinical and pathophysiological aspects.Acta Myol. 2005 Oct;24(2):104-9. Acta Myol. 2005. PMID: 16550926 Review.
Cited by
-
Deletion of MLIP (muscle-enriched A-type lamin-interacting protein) leads to cardiac hyperactivation of Akt/mammalian target of rapamycin (mTOR) and impaired cardiac adaptation.J Biol Chem. 2015 Oct 30;290(44):26699-714. doi: 10.1074/jbc.M115.678433. Epub 2015 Sep 10. J Biol Chem. 2015. PMID: 26359501 Free PMC article.
-
Native lamin A/C proteomes and novel partners from heart and skeletal muscle in a mouse chronic inflammation model of human frailty.Front Cell Dev Biol. 2023 Oct 23;11:1240285. doi: 10.3389/fcell.2023.1240285. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37936983 Free PMC article.
-
Nuclear membrane diversity: underlying tissue-specific pathologies in disease?Curr Opin Cell Biol. 2015 Jun;34:101-12. doi: 10.1016/j.ceb.2015.06.003. Epub 2015 Jun 24. Curr Opin Cell Biol. 2015. PMID: 26115475 Free PMC article. Review.
-
Lamins at a glance.J Cell Sci. 2012 May 1;125(Pt 9):2087-93. doi: 10.1242/jcs.087288. J Cell Sci. 2012. PMID: 22669459 Free PMC article. Review. No abstract available.
-
Broken nuclei--lamins, nuclear mechanics, and disease.Trends Cell Biol. 2014 Apr;24(4):247-56. doi: 10.1016/j.tcb.2013.11.004. Epub 2013 Dec 2. Trends Cell Biol. 2014. PMID: 24309562 Free PMC article. Review.
References
-
- Cohen M., Lee K. K., Wilson K. L., Gruenbaum Y. (2001) Trends Biochem. Sci. 26, 41–47 - PubMed
-
- Mounkes L. C., Kozlov S., Hernandez L., Sullivan T., Stewart C. L. (2003) Nature 423, 298–301 - PubMed
-
- Röber R. A., Weber K., Osborn M. (1989) Development 105, 365–378 - PubMed
-
- Constantinescu D., Gray H. L., Sammak P. J., Schatten G. P., Csoka A. B. (2006) Stem Cells 24, 177–185 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

