How does DNA break during chromosomal translocations?
- PMID: 21498543
- PMCID: PMC3152359
- DOI: 10.1093/nar/gkr223
How does DNA break during chromosomal translocations?
Abstract
Chromosomal translocations are one of the most common types of genetic rearrangements and are molecular signatures for many types of cancers. They are considered as primary causes for cancers, especially lymphoma and leukemia. Although many translocations have been reported in the last four decades, the mechanism by which chromosomes break during a translocation remains largely unknown. In this review, we summarize recent advances made in understanding the molecular mechanism of chromosomal translocations.
Figures
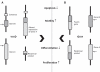
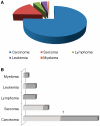
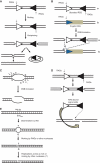
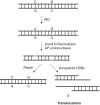
References
-
- Boveri T. Zur Frage der Entstehung maligner Tumoren [The Origin of Malignant Tumors] 1914. Gustav Fischer, Jena.
-
- Stern C. Boveri and the early days of genetics. Nature. 1950;166:446. - PubMed
-
- Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497. - PubMed
-
- Rowley JD. A new consistent chromosomal abnormality in chronic Myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. - PubMed
-
- Zech L, Haglund U, Nilsson K, Klein G. Characteristic chromosomal abnormalities in biopsies and lymphoid-cell lines from patients with Burkitt and non-Burkitt lymphomas. Int. J. Cancer. 1976;17:47–56. - PubMed

