Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells
- PMID: 21498568
- PMCID: PMC3078704
- DOI: 10.1101/gad.2027911
Polycomb EZH2 controls self-renewal and safeguards the transcriptional identity of skeletal muscle stem cells
Abstract
Satellite cells (SCs) sustain muscle growth and empower adult skeletal muscle with vigorous regenerative abilities. Here, we report that EZH2, the enzymatic subunit of the Polycomb-repressive complex 2 (PRC2), is expressed in both Pax7+/Myf5⁻ stem cells and Pax7+/Myf5+ committed myogenic precursors and is required for homeostasis of the adult SC pool. Mice with conditional ablation of Ezh2 in SCs have fewer muscle postnatal Pax7+ cells and reduced muscle mass and fail to appropriately regenerate. These defects are associated with impaired SC proliferation and derepression of genes expressed in nonmuscle cell lineages. Thus, EZH2 controls self-renewal and proliferation, and maintains an appropriate transcriptional program in SCs.
Figures
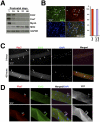
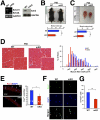
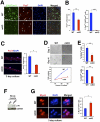
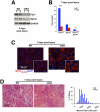

Comment in
-
Polycomb-mediated repression during terminal differentiation: what don't you want to be when you grow up?Genes Dev. 2011 May 15;25(10):997-1003. doi: 10.1101/gad.2054311. Genes Dev. 2011. PMID: 21576260 Free PMC article.
References
-
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 - PubMed
-
- Buckingham M, Relaix F 2007. The role of Pax genes in the development of tissues and organs: Pax3 and Pax7 regulate muscle progenitor cell functions. Annu Rev Cell Dev Biol 23: 645–673 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
