A kinase shRNA screen links LATS2 and the pRB tumor suppressor
- PMID: 21498571
- PMCID: PMC3078707
- DOI: 10.1101/gad.2000211
A kinase shRNA screen links LATS2 and the pRB tumor suppressor
Abstract
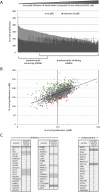
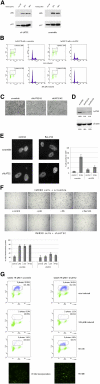
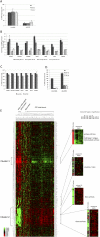
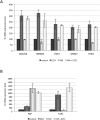
pRB-mediated inhibition of cell proliferation is a complex process that depends on the action of many proteins. However, little is known about the specific pathways that cooperate with the Retinoblastoma protein (pRB) and the variables that influence pRB's ability to arrest tumor cells. Here we describe two shRNA screens that identify kinases that are important for pRB to suppress cell proliferation and pRB-mediated induction of senescence markers. The results reveal an unexpected effect of LATS2, a component of the Hippo pathway, on pRB-induced phenotypes. Partial knockdown of LATS2 strongly suppresses some pRB-induced senescence markers. Further analysis shows that LATS2 cooperates with pRB to promote the silencing of E2F target genes, and that reduced levels of LATS2 lead to defects in the assembly of DREAM (DP, RB [retinoblastoma], E2F, and MuvB) repressor complexes at E2F-regulated promoters. Kinase assays show that LATS2 can phosphorylate DYRK1A, and that it enhances the ability of DYRK1A to phosphorylate the DREAM subunit LIN52. Intriguingly, the LATS2 locus is physically linked with RB1 on 13q, and this region frequently displays loss of heterozygosity in human cancers. Our results reveal a functional connection between the pRB and Hippo tumor suppressor pathways, and suggest that low levels of LATS2 may undermine the ability of pRB to induce a permanent cell cycle arrest in tumor cells.
Figures


Comment in
-
Sweet DREAMs for Hippo.Genes Dev. 2011 May 1;25(9):889-94. doi: 10.1101/gad.2050411. Genes Dev. 2011. PMID: 21536729 Free PMC article.
References
-
- Adams PD 2009. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol Cell 36: 2–14 - PubMed
-
- Alexander K, Yang HS, Hinds PW 2003. pRb inactivation in senescent cells leads to an E2F-dependent apoptosis requiring p73. Mol Cancer Res 1: 716–728 - PubMed
-
- Alonso A, Rahmouni S, Williams S, van Stipdonk M, Jaroszewski L, Godzik A, Abraham RT, Schoenberger SP, Mustelin T 2003. Tyrosine phosphorylation of VHR phosphatase by ZAP-70. Nat Immunol 4: 44–48 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
