Extensive DNA-binding specificity divergence of a conserved transcription regulator
- PMID: 21498688
- PMCID: PMC3088634
- DOI: 10.1073/pnas.1019177108
Extensive DNA-binding specificity divergence of a conserved transcription regulator
Abstract
The DNA sequence recognized by a transcription regulator can be conserved across large evolutionary distances. For example, it is known that many homologous regulators in yeasts and mammals can recognize the same (or closely related) DNA sequences. In contrast to this paradigm, we describe a case in which the DNA-binding specificity of a transcription regulator has changed so extensively (and over a much smaller evolutionary distance) that its cis-regulatory sequence appears unrelated in different species. Bioinformatic, genetic, and biochemical approaches were used to document and analyze a major change in the DNA-binding specificity of Matα1, a regulator of cell-type specification in ascomycete fungi. Despite this change, Matα1 controls the same core set of genes in the hemiascomycetes because its DNA recognition site has evolved with it, preserving the protein-DNA interaction but significantly changing its molecular details. Matα1 and its recognition sequence diverged most dramatically in the common ancestor of the CTG-clade (Candida albicans, Candida lusitaniae, and related species), apparently without the aid of a gene duplication event. Our findings suggest that DNA-binding specificity divergence between orthologous transcription regulators may be more prevalent than previously thought and that seemingly unrelated cis-regulatory sequences can nonetheless be homologous. These findings have important implications for understanding transcriptional network evolution and for the bioinformatic analysis of regulatory circuits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
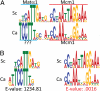
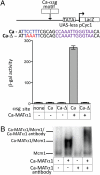


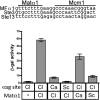
Similar articles
-
Conservation and evolution of cis-regulatory systems in ascomycete fungi.PLoS Biol. 2004 Dec;2(12):e398. doi: 10.1371/journal.pbio.0020398. Epub 2004 Nov 9. PLoS Biol. 2004. PMID: 15534694 Free PMC article.
-
Intercalation of a new tier of transcription regulation into an ancient circuit.Nature. 2010 Dec 16;468(7326):959-63. doi: 10.1038/nature09560. Nature. 2010. PMID: 21164485 Free PMC article.
-
Engineered improvements in DNA-binding function of the MATa1 homeodomain reveal structural changes involved in combinatorial control.J Mol Biol. 2002 Feb 15;316(2):247-56. doi: 10.1006/jmbi.2001.5333. J Mol Biol. 2002. PMID: 11851335
-
From elements to modules: regulatory evolution in Ascomycota fungi.Curr Opin Genet Dev. 2009 Dec;19(6):571-8. doi: 10.1016/j.gde.2009.09.007. Epub 2009 Oct 29. Curr Opin Genet Dev. 2009. PMID: 19879128 Free PMC article. Review.
-
Evolutionary Transition of GAL Regulatory Circuit from Generalist to Specialist Function in Ascomycetes.Trends Microbiol. 2018 Aug;26(8):692-702. doi: 10.1016/j.tim.2017.12.008. Trends Microbiol. 2018. PMID: 29395731 Review.
Cited by
-
Making sense of transcription networks.Cell. 2015 May 7;161(4):714-23. doi: 10.1016/j.cell.2015.04.014. Cell. 2015. PMID: 25957680 Free PMC article. Review.
-
Interplay of Chimeric Mating-Type Loci Impairs Fertility Rescue and Accounts for Intra-Strain Variability in Zygosaccharomyces rouxii Interspecies Hybrid ATCC42981.Front Genet. 2019 Mar 1;10:137. doi: 10.3389/fgene.2019.00137. eCollection 2019. Front Genet. 2019. PMID: 30881382 Free PMC article.
-
Evolutionary divergence of the sex-determining gene MID uncoupled from the transition to anisogamy in volvocine algae.Development. 2018 Apr 9;145(7):dev162537. doi: 10.1242/dev.162537. Development. 2018. PMID: 29549112 Free PMC article.
-
High-Throughput Identification of Cis-Regulatory Rewiring Events in Yeast.Mol Biol Evol. 2015 Dec;32(12):3047-63. doi: 10.1093/molbev/msv203. Epub 2015 Sep 23. Mol Biol Evol. 2015. PMID: 26399482 Free PMC article.
-
Evolutionary principles of modular gene regulation in yeasts.Elife. 2013 Jun 18;2:e00603. doi: 10.7554/eLife.00603. Elife. 2013. PMID: 23795289 Free PMC article.
References
-
- Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. - PubMed
-
- McGinnis N, Kuziora MA, McGinnis W. Human Hox-4.2 and Drosophila deformed encode similar regulatory specificities in Drosophila embryos and larvae. Cell. 1990;63:969–976. - PubMed
-
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

