Genome-wide transcriptional and physiological responses of Bradyrhizobium japonicum to paraquat-mediated oxidative stress
- PMID: 21498770
- PMCID: PMC3127611
- DOI: 10.1128/AEM.00047-11
Genome-wide transcriptional and physiological responses of Bradyrhizobium japonicum to paraquat-mediated oxidative stress
Abstract
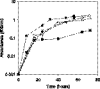
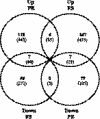
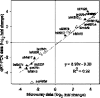
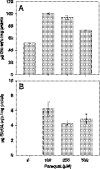
The rhizobial bacterium Bradyrhizobium japonicum functions as a nitrogen-fixing symbiont of the soybean plant (Glycine max). Plants are capable of producing an oxidative burst, a rapid proliferation of reactive oxygen species (ROS), as a defense mechanism against pathogenic and symbiotic bacteria. Therefore, B. japonicum must be able to resist such a defense mechanism to initiate nodulation. In this study, paraquat, a known superoxide radical-inducing agent, was used to investigate this response. Genome-wide transcriptional profiles were created for both prolonged exposure (PE) and fulminant shock (FS) conditions. These profiles revealed that 190 and 86 genes were up- and downregulated for the former condition, and that 299 and 105 genes were up- and downregulated for the latter condition, respectively (>2.0-fold; P < 0.05). Many genes within putative operons for F(0)F(1)-ATP synthase, chemotaxis, transport, and ribosomal proteins were upregulated during PE. The transcriptional profile for the FS condition strangely resembled that of a bacteroid condition, including the FixK(2) transcription factor and most of its response elements. However, genes encoding canonical ROS scavenging enzymes, such as superoxide dismutase and catalase, were not detected, suggesting constitutive expression of those genes by endogenous ROS. Various physiological tests, including exopolysaccharide (EPS), cellular protein, and motility characterization, were performed to corroborate the gene expression data. The results suggest that B. japonicum responds to tolerable oxidative stress during PE through enhanced motility, increased translational activity, and EPS production, in addition to the expression of genes involved in global stress responses, such as chaperones and sigma factors.
Figures




References
-
- Adler J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77–91 - PubMed
-
- Baker C. J., Orlandi E. W. 1995. Active oxygen in plant pathogenesis. Annu. Rev. Phytopathol. 33:299–321 - PubMed
-
- Baker C. J., Mock N., Glazener J., Orlandi E. 1993. Recognition responses in pathogen/non-host and race/cultivar interactions involving soybean (Glycine max) and Pseudomonas syringae pathovars. Physiol. Mol. Plant Pathol. 43:81–94
-
- Barloy-Hubler F., Cheron A., Hellegouarch A., Galibert F. 2004. Smc01944, a secreted peroxidase induced by oxidative stresses in Sinorhizobium meliloti 1021. Microbiology 150:657–664 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

