Faster than neutral evolution of constrained sequences: the complex interplay of mutational biases and weak selection
- PMID: 21498884
- PMCID: PMC3101017
- DOI: 10.1093/gbe/evr032
Faster than neutral evolution of constrained sequences: the complex interplay of mutational biases and weak selection
Abstract
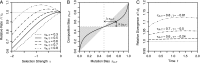
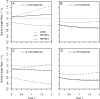
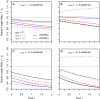
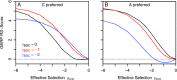
Comparative genomics has become widely accepted as the major framework for the ascertainment of functionally important regions in genomes. The underlying paradigm of this approach is that most of the functional regions are assumed to be under selective constraint, which in turn reduces the rate of evolution relative to neutrality. This assumption allows detection of functional regions through sequence conservation. However, constraint does not always lead to sequence conservation. When purifying selection is weak and mutation is biased, constrained regions can even evolve faster than neutral sequences and thus can appear to be under positive selection. Moreover, conservation estimates depend also on the orientation of selection relative to mutational biases and can vary over time. In the light of recent data of the ubiquity of mutational biases and weak selective forces, these effects should reduce the power of conservation analyses to define functional regions using comparative genomics data. We argue that the estimation of true mutational biases and the use of explicit evolutionary models are essential to improve methods inferring the action of natural selection and functionality in genome sequences.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

