NFIL3/E4BP4 controls type 2 T helper cell cytokine expression
- PMID: 21499227
- PMCID: PMC3098483
- DOI: 10.1038/emboj.2011.111
NFIL3/E4BP4 controls type 2 T helper cell cytokine expression
Abstract
Type 2 T helper (T(H)2) cells are critical for the development of allergic immune responses; however, the molecular mechanism controlling their effector function is still largely unclear. Here, we report that the transcription factor NFIL3/E4BP4 regulates cytokine production and effector function by T(H)2 cells. NFIL3 is highly expressed in T(H)2 cells but much less in T(H)1 cells. Production of interleukin (IL)-13 and IL-5 is significantly increased in Nfil3(-/-) T(H)2 cells and is decreased by expression of NFIL3 in wild-type T(H)2 cells. NFIL3 directly binds to and negatively regulates the Il13 gene. In contrast, IL-4 production is decreased in Nfil3(-/-) T(H)2 cells. Increased IL-13 and IL-5 together with decreased IL-4 production by antigen-stimulated splenocytes from the immunized Nfil3(-/-) mice was also observed. The ability of NFIL3 to alter T(H)2 cytokine production is a T-cell intrinsic effect. Taken together, these data indicate that NFIL3 is a key regulator of T(H)2 responses.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

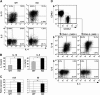

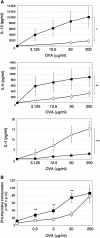


Similar articles
-
The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells.Nat Immunol. 2011 May;12(5):450-9. doi: 10.1038/ni.2020. Epub 2011 Apr 3. Nat Immunol. 2011. PMID: 21460847 Free PMC article.
-
IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching.Proc Natl Acad Sci U S A. 2010 Jan 12;107(2):821-6. doi: 10.1073/pnas.0909235107. Epub 2009 Dec 22. Proc Natl Acad Sci U S A. 2010. PMID: 20080759 Free PMC article.
-
α1-adrenergic receptor signaling in osteoblasts regulates clock genes and bone morphogenetic protein 4 expression through up-regulation of the transcriptional factor nuclear factor IL-3 (Nfil3)/E4 promoter-binding protein 4 (E4BP4).J Biol Chem. 2014 Jun 13;289(24):17174-83. doi: 10.1074/jbc.M113.546135. Epub 2014 May 2. J Biol Chem. 2014. PMID: 24794868 Free PMC article.
-
The transcriptional regulator NFIL3 controls IgE production.Trans Am Clin Climatol Assoc. 2010;121:156-71; discussion 171. Trans Am Clin Climatol Assoc. 2010. PMID: 20697558 Free PMC article. Review.
-
Regulation of Th2 cytokine gene transcription.Chem Immunol. 2000;78:16-29. doi: 10.1159/000058813. Chem Immunol. 2000. PMID: 12847715 Review.
Cited by
-
Bach2 maintains T cells in a naive state by suppressing effector memory-related genes.Proc Natl Acad Sci U S A. 2013 Jun 25;110(26):10735-40. doi: 10.1073/pnas.1306691110. Epub 2013 Jun 10. Proc Natl Acad Sci U S A. 2013. PMID: 23754397 Free PMC article.
-
The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development.J Exp Med. 2015 Jun 1;212(6):865-74. doi: 10.1084/jem.20142318. Epub 2015 May 11. J Exp Med. 2015. PMID: 25964371 Free PMC article.
-
Identifying hub circadian rhythm biomarkers and immune cell infiltration in rheumatoid arthritis.Front Immunol. 2022 Sep 27;13:1004883. doi: 10.3389/fimmu.2022.1004883. eCollection 2022. Front Immunol. 2022. PMID: 36238290 Free PMC article.
-
Glucocorticoid circadian rhythms in immune function.Semin Immunopathol. 2022 Mar;44(2):153-163. doi: 10.1007/s00281-021-00889-2. Epub 2021 Sep 28. Semin Immunopathol. 2022. PMID: 34580744 Review.
-
Antigen exposure in the late light period induces severe symptoms of food allergy in an OVA-allergic mouse model.Sci Rep. 2015 Sep 30;5:14424. doi: 10.1038/srep14424. Sci Rep. 2015. PMID: 26419283 Free PMC article.
References
-
- Agarwal S, Rao A (1998) Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9: 765–775 - PubMed
-
- Altura RA, Inukai T, Ashmun RA, Zambetti GP, Roussel MF, Look AT (1998) The chimeric E2A-HLF transcription factor abrogates p53-induced apoptosis in myeloid leukemia cells. Blood 92: 1397–1405 - PubMed
-
- Ansel KM, Djuretic I, Tanasa B, Rao A (2006) Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol 24: 607–656 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

