Cryo-EM structure of the ribosome-SecYE complex in the membrane environment
- PMID: 21499241
- PMCID: PMC3412285
- DOI: 10.1038/nsmb.2026
Cryo-EM structure of the ribosome-SecYE complex in the membrane environment
Abstract
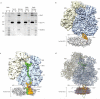
The ubiquitous SecY-Sec61 complex translocates nascent secretory proteins across cellular membranes and integrates membrane proteins into lipid bilayers. Several structures of mostly detergent-solubilized Sec complexes have been reported. Here we present a single-particle cryo-EM structure of the SecYEG complex in a membrane environment, bound to a translating ribosome, at subnanometer resolution. Using the SecYEG complex reconstituted in a so-called Nanodisc, we could trace the nascent polypeptide chain from the peptidyltransferase center into the membrane. The reconstruction allowed for the identification of ribosome-lipid interactions. The rRNA helix 59 (H59) directly contacts the lipid surface and appears to modulate the membrane in immediate vicinity to the proposed lateral gate of the protein-conducting channel (PCC). On the basis of our map and molecular dynamics simulations, we present a model of a signal anchor-gated PCC in the membrane.
Figures
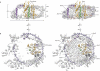

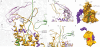

References
-
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450(7170):663–669. - PubMed
-
- Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol. 2005;15(1):116–125. - PubMed
-
- Van den Berg B, et al. X–ray structure of a protein–conducting channel. Nature. 2004;427(6969):36–44. Epub 2003 Dec 2003. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

