Molecular targeting of CSN5 in human hepatocellular carcinoma: a mechanism of therapeutic response
- PMID: 21499307
- PMCID: PMC3140552
- DOI: 10.1038/onc.2011.126
Molecular targeting of CSN5 in human hepatocellular carcinoma: a mechanism of therapeutic response
Abstract
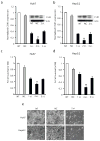
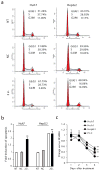
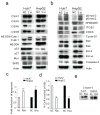
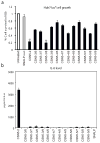
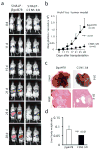
Development of targeted therapy for hepatocellular carcinoma (HCC) remains a major challenge. We have recently identified an elevated expression of the fifth subunit of COP9 signalosome (CSN5) in early HCC as compared with dysplastic stage. In the present study, we explored the possibility of CSN5 being a potential therapeutic target for HCC. Our results show that CSN5 knockdown by small-interfering (si) RNA caused a strong induction of apoptosis and inhibition of cell-cycle progression in HCC cells in vitro. The down-regulation of CSN5 was sufficient to interfere with CSN function as evidenced by the accumulation of neddylated Cullin 1 and changes in the protein levels of CSN-controlled substrates SKP2, p53, p27 and nuclear factor-κB, albeit to a different degree depending on the HCC cell line, which could account for the CSN5 knockdown phenotype. The transcriptomic analysis of CSN5 knockdown signature showed that the anti-proliferative effect was driven by a common subset of molecular alterations including down-regulation of cyclin-dependent kinase 6 (CDK6) and integrin β1 (ITGB1), which were functionally interconnected with key oncogenic regulators MYC and TGFβ1 involved in the control of proliferation, apoptotic cell death and HCC progression. Consistent with microarray analysis, western blotting revealed that CSN5 depletion increased phosphorylation of Smad 2/3, key mediators of TGFβ1 signaling, decreased the protein levels of ITGB1, CDK6 and cyclin D1 and caused reduced expression of anti-apoptotic Bcl-2, while elevating the levels of pro-apoptotic Bak. A chemically modified variant of CSN5 siRNA was then selected for in vivo application based on the growth inhibitory effect and minimal induction of unwanted immune response. Systemic delivery of the CSN5 3/8 variant by stable-nucleic-acid-lipid particles significantly suppressed the tumor growth in Huh7-luc+ orthotopic xenograft model. Taken together, these results indicate that CSN5 has a pivotal role in HCC pathogenesis and maybe an attractive molecular target for systemic HCC therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Arsura M, Cavin LG. Nuclear factor-kappaB and liver carcinogenesis. Cancer Lett. 2005;229:157–169. - PubMed
-
- Avila MA, Berasain C, Torres L, Martín-Duce A, Corrales FJ, Yang H, et al. Reduced mRNA abundance of the main enzymes involved in methionine metabolism in human liver cirrhosis and hepatocellular carcinoma. J Hepatol. 2000;33:907–914. - PubMed
-
- Bae MK, Ahn MY, Jeong JW, Bae MH, Lee YM, Bae SK, et al. Jab1 interacts directly with HIF-1alpha and regulates its stability. J Biol Chem. 2002;277:9–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

