Cell-based Models for Discovery of Pharmacogenomic Markers of Anticancer Agent Toxicity
- PMID: 21499559
- PMCID: PMC3076057
Cell-based Models for Discovery of Pharmacogenomic Markers of Anticancer Agent Toxicity
Abstract
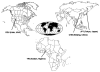
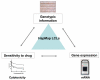
The field of pharmacogenomics is challenging because of the multigenic nature of drug response and toxicity. The candidate gene approach has been traditionally utilized to determine the contribution of genetic variation to a particular phenotype; however, the sequencing of the human genome and the genetic resource provided by the International HapMap Project has allowed researchers to perform genome-wide studies without a priori knowledge. Recent work has demonstrated the usefulness of cell-based models for pharmacogenomic discovery using the HapMap samples, which are a panel of well-genotyped, human lymphoblastoid cell lines (LCLs) derived from 90 Utah residents with ancestry from northern and western Europe (CEU), 90 Yoruba in Ibadan, Nigeria (YRI), 45 Japanese in Tokyo, Japan (JPT) and 45 Han Chinese in Beijing, China (CHB). Using these cell-based models, investigators are able to study not only individual variation in drug response, but also population differences in drug response. Finally, besides single nucleotide polymorphisms (SNPs) and gene expression, these cell-based models can also be used to investigate other genetic (e.g. copy number variants, CNVs), epigenetic or environmental factors responsible for drug response.
Figures
Similar articles
-
Genetic polymorphisms of pharmacogenomic VIP variants in the Kyrgyz population from northwest China.Gene. 2013 Oct 15;529(1):88-93. doi: 10.1016/j.gene.2013.07.078. Epub 2013 Aug 13. Gene. 2013. PMID: 23954225
-
[A cross-racial analysis on the susceptible gene polymorphisms of salt-sensitive hypertension].Zhonghua Xin Xue Guan Bing Za Zhi. 2010 Oct;38(10):943-8. Zhonghua Xin Xue Guan Bing Za Zhi. 2010. PMID: 21176642 Chinese.
-
[Analysis of the HapMap data on SNPs in SUMO1 and association study of rs7599810 in trios with non-syndromic cleft lip with or without cleft palate].Beijing Da Xue Xue Bao Yi Xue Ban. 2014 Apr 18;46(2):258-63. Beijing Da Xue Xue Bao Yi Xue Ban. 2014. PMID: 24743817 Chinese.
-
Pharmacogenomics of chemotherapeutic susceptibility and toxicity.Genome Med. 2012 Nov 30;4(11):90. doi: 10.1186/gm391. eCollection 2012. Genome Med. 2012. PMID: 23199206 Free PMC article. Review.
-
Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation.Pharmacogenomics. 2012 Jan;13(1):55-70. doi: 10.2217/pgs.11.121. Pharmacogenomics. 2012. PMID: 22176622 Free PMC article. Review.
Cited by
-
A pharmacogene database enhanced by the 1000 Genomes Project.Pharmacogenet Genomics. 2009 Oct;19(10):829-32. doi: 10.1097/FPC.0b013e3283317bac. Pharmacogenet Genomics. 2009. PMID: 19745786 Free PMC article.
-
Utility of patient-derived lymphoblastoid cell lines as an ex vivo capecitabine sensitivity prediction model for breast cancer patients.Oncotarget. 2016 Jun 21;7(25):38359-38366. doi: 10.18632/oncotarget.9521. Oncotarget. 2016. PMID: 27224917 Free PMC article.
-
Gene expression changes in lymphoblastoid cell lines and primary B cells by dexamethasone.Pharmacogenet Genomics. 2019 Apr;29(3):58-64. doi: 10.1097/FPC.0000000000000365. Pharmacogenet Genomics. 2019. PMID: 30562215 Free PMC article.
-
A comparison of association methods for cytotoxicity mapping in pharmacogenomics.Front Genet. 2011 Dec 14;2:86. doi: 10.3389/fgene.2011.00086. eCollection 2011. Front Genet. 2011. PMID: 22303380 Free PMC article.
-
Integrating cell-based and clinical genome-wide studies to identify genetic variants contributing to treatment failure in neuroblastoma patients.Clin Pharmacol Ther. 2014 Jun;95(6):644-52. doi: 10.1038/clpt.2014.37. Epub 2014 Feb 18. Clin Pharmacol Ther. 2014. PMID: 24549002 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
