The Nucleolus Takes Control of Protein Trafficking Under Cellular Stress
- PMID: 21499571
- PMCID: PMC3076688
The Nucleolus Takes Control of Protein Trafficking Under Cellular Stress
Abstract
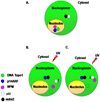
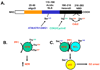
The nucleolus is a highly dynamic nuclear substructure that was originally described as the site of ribosome biogenesis. The advent of proteomic analysis has now allowed the identification of over 4500 nucleolus associated proteins with only about 30% of them associated with ribogenesis (1). The great number of nucleolar proteins not associated with traditionally accepted nucleolar functions indicates a role for the nucleolus in other cellular functions such as mitosis, cell-cycle progression, cell proliferation and many forms of stress response including DNA repair (2). A number of recent reviews have addressed the pivotal role of the nucleolus in the cellular stress response (1, 3, 4). Here, we will focus on the role of Nucleolin and Nucleophosmin, two major components of the nucleolus, in response to genotoxic stress. Due to space constraint only a limited number of studies are cited. We thus apologize to all our colleagues whose works are not referenced here.
Conflict of interest statement
No potential conflicts of interest to disclose.
Figures

Similar articles
-
Nucleophosmin: A Nucleolar Phosphoprotein Orchestrating Cellular Stress Responses.Cells. 2024 Jul 27;13(15):1266. doi: 10.3390/cells13151266. Cells. 2024. PMID: 39120297 Free PMC article. Review.
-
Lamin B2 Modulates Nucleolar Morphology, Dynamics, and Function.Mol Cell Biol. 2017 Nov 28;37(24):e00274-17. doi: 10.1128/MCB.00274-17. Print 2017 Dec 15. Mol Cell Biol. 2017. PMID: 28993479 Free PMC article.
-
Nucleolin and nucleophosmin: nucleolar proteins with multiple functions in DNA repair.Biochem Cell Biol. 2016 Oct;94(5):419-432. doi: 10.1139/bcb-2016-0068. Epub 2016 Jun 29. Biochem Cell Biol. 2016. PMID: 27673355 Review.
-
The role of the nucleolus in regulating the cell cycle and the DNA damage response.Adv Protein Chem Struct Biol. 2023;135:203-241. doi: 10.1016/bs.apcsb.2023.01.001. Epub 2023 Feb 8. Adv Protein Chem Struct Biol. 2023. PMID: 37061332 Review.
-
Nucleolar protein trafficking in response to HIV-1 Tat: rewiring the nucleolus.PLoS One. 2012;7(11):e48702. doi: 10.1371/journal.pone.0048702. Epub 2012 Nov 15. PLoS One. 2012. PMID: 23166591 Free PMC article.
Cited by
-
Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells.Mol Biol Cell. 2012 Oct;23(20):4079-96. doi: 10.1091/mbc.E12-04-0299. Epub 2012 Aug 23. Mol Biol Cell. 2012. PMID: 22918947 Free PMC article.
-
Nucleolar stress in C9orf72 and sporadic ALS spinal motor neurons precedes TDP-43 mislocalization.Acta Neuropathol Commun. 2021 Feb 15;9(1):26. doi: 10.1186/s40478-021-01125-6. Acta Neuropathol Commun. 2021. PMID: 33588953 Free PMC article.
-
Nucleophosmin: A Nucleolar Phosphoprotein Orchestrating Cellular Stress Responses.Cells. 2024 Jul 27;13(15):1266. doi: 10.3390/cells13151266. Cells. 2024. PMID: 39120297 Free PMC article. Review.
-
C1D family proteins in coordinating RNA processing, chromosome condensation and DNA damage response.Cell Div. 2016 Mar 9;11:2. doi: 10.1186/s13008-016-0014-5. eCollection 2016. Cell Div. 2016. PMID: 27030795 Free PMC article. Review.
-
Heat-shock stress activates a novel nuclear import pathway mediated by Hikeshi.Nucleus. 2012 Sep-Oct;3(5):422-8. doi: 10.4161/nucl.21713. Epub 2012 Aug 16. Nucleus. 2012. PMID: 22895094 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
